2K25
 
 | |
2K22
 
 | |
2K24
 
 | |
1PES
 
 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
1PET
 
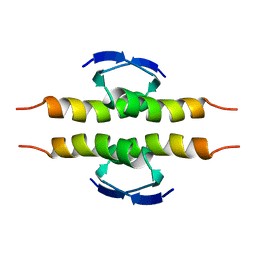 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
1VNA
 
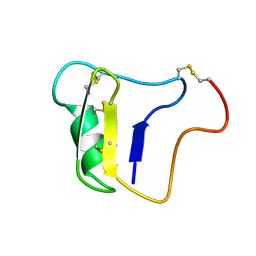 | |
1VNB
 
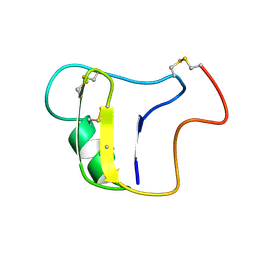 | |
2M5T
 
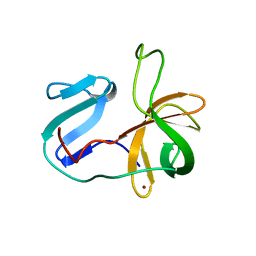 | | Solution structure of the 2A proteinase from a common cold agent, human rhinovirus RV-C02, strain W12 | | Descriptor: | ZINC ION, human rhinovirus 2A proteinase | | Authors: | Lee, W, Frederick, R, Tonelli, M, Troupis, A.T, Reinin, N, Suchy, F.P, Moyer, K, Watters, K, Aceti, D, Palmenberg, A.C, Markley, J.L. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 2A Protease from a Common Cold Agent, Human Rhinovirus C2, Strain W12.
Plos One, 9, 2014
|
|
2KGT
 
 | |
7KNV
 
 | | Solution NMR structure of CDHR3 extracellular domain EC1 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3 | | Authors: | Lee, W, Tonelli, M, Frederick, R.O, Watters, K.E, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2020-11-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Determination of the CDHR3 Rhinovirus-C Binding Domain, EC1
Viruses, 13, 2021
|
|
7LU8
 
 | |
6NU4
 
 | | Solution structure of the Arabidopsis thaliana RALF8 peptide | | Descriptor: | Protein RALF-like 8 | | Authors: | Lee, W, Markley, J.L, Frederick, R.O, Miyoshi, H, Tonelli, M, Cornilescu, G, Cornilescu, C, Sussman, M.R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Function and solution structure of the Arabidopsis thaliana RALF8 peptide.
Protein Sci., 28, 2019
|
|
6LZP
 
 | |
2ROH
 
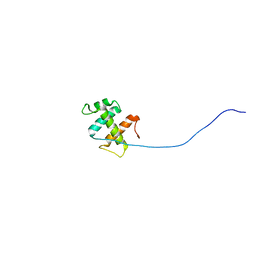 | | The DNA binding domain of RTBP1 | | Descriptor: | Telomere binding protein-1 | | Authors: | Lee, W, Ko, S. | | Deposit date: | 2008-03-22 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of rice telomere binding protein RTBP1
Biochemistry, 48, 2009
|
|
6KZ7
 
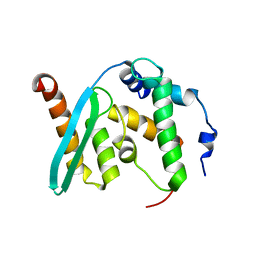 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | Descriptor: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | Deposit date: | 2019-09-23 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
2M17
 
 | |
1HVB
 
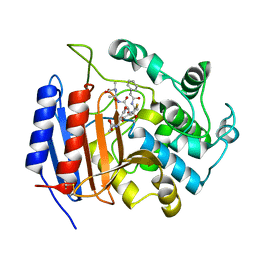 | | CRYSTAL STRUCTURE OF STREPTOMYCES R61 DD-PEPTIDASE COMPLEXED WITH A NOVEL CEPHALOSPORIN ANALOG OF CELL WALL PEPTIDOGLYCAN | | Descriptor: | 5-{3-(S)-(4-(R)-ACETYLAMINO-4-CARBOXY-BUTYRYLAMINO)-3-[1-(R)-(1-(R)-CARBOXY-ETHYLCARBAMOYL)-ETHYLCARBAMOYL]-PROPYL}-2-( CARBOXY-PHENYLACETYLAMINO-METHYL)-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE | | Authors: | McDonough, M.A, Lee, W, Silvaggi, N.R, Mobashery, S, Kelly, J.A. | | Deposit date: | 2001-01-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A 1.2-A snapshot of the final step of bacterial cell wall biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2KX2
 
 | |
7LHQ
 
 | | Solution structure of SARS-CoV-2 nonstructural protein 7 at pH 7.0 | | Descriptor: | Non-structural protein 7 | | Authors: | Lee, Y, Tonelli, M, Anderson, T.K, Kirchdoerfer, R.N, Henzler-Wildman, K, Lee, W. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-dependent polymorphism of the structure of SARS-CoV-2 nsp7
Biorxiv, 2021
|
|
1R02
 
 | |
2ABY
 
 | | Solution structure of TA0743 from Thermoplasma acidophilum | | Descriptor: | hypothetical protein TA0743 | | Authors: | Kim, B, Jung, J, Hong, E, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2005-07-18 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the conserved novel-fold protein TA0743 from Thermoplasma acidophilum.
Proteins, 62, 2006
|
|
8Y4Z
 
 | | Monomeric HERC5 HECT c-lobe structure in solution | | Descriptor: | E3 ISG15--protein ligase HERC5 | | Authors: | Dag, C, Lambert, M, Kahraman, K, Lohn, F, Lee, W, Gocenler, O, Guntert, P, Dotsch, V. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Monomeric HERC5 HECT c-lobe structure in solution
To Be Published
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | Descriptor: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | Descriptor: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
