5MLH
 
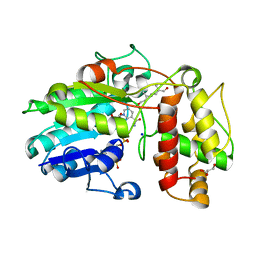 | | Plantago Major multifunctional oxidoreductase in complex with 8-oxogeranial and NADP+ | | Descriptor: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, CALCIUM ION, GLYCEROL, ... | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-06 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
5MLR
 
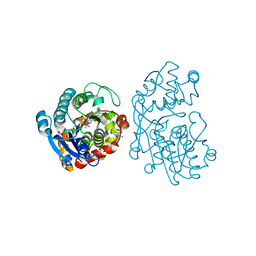 | | Plantago Major multifunctional oxidoreductase V150M mutant in complex with citral and NADP+ | | Descriptor: | CHLORIDE ION, Geranaldehyde, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
4ZZ7
 
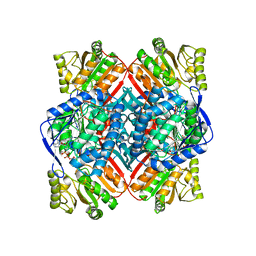 | | Crystal structure of methylmalonate-semialdehyde dehydrogenase (DddC) from Oceanimonas doudoroffii | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Do, H, Lee, C.W, Lee, S.G, Kang, H, Park, C.M, Kim, H.J, Park, H, Park, H, Lee, J.H. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii.
J. Microbiol., 54, 2016
|
|
7DH0
 
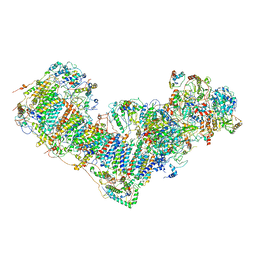 | | Activity optimized complex I (open form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGZ
 
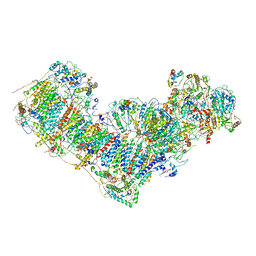 | | Activity optimized complex I (closed form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGS
 
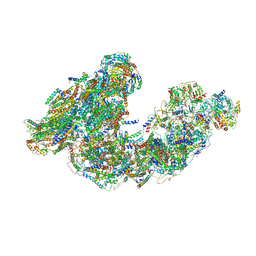 | | Activity optimized supercomplex state3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGR
 
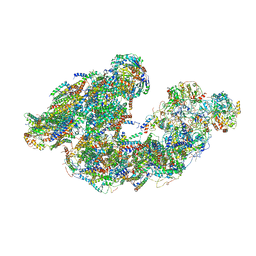 | | Activity optimized supercomplex state2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGQ
 
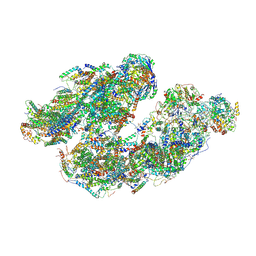 | | Activity optimized supercomplex state1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DKF
 
 | | Activity optimized supercomplex state4 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
5YSS
 
 | |
6LT4
 
 | | AAA+ ATPase, ClpL from Streptococcus pneumoniae: ATPrS-bound | | Descriptor: | ATP-dependent Clp protease, ATP-binding subunit, MAGNESIUM ION, ... | | Authors: | Kim, G, Lee, S.G, Han, S, Jung, J, Jeong, H.S, Hyun, J.K, Rhee, D.K, Kim, H.M, Lee, S. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | ClpL is a functionally active tetradecameric AAA+ chaperone, distinct from hexameric/dodecameric ones.
Faseb J., 34, 2020
|
|
6LSY
 
 | | AAA+ ATPase, ClpL from Streptococcus pneumoniae - ATP bound | | Descriptor: | ATP-dependent Clp protease, ATP-binding subunit | | Authors: | Kim, G, Lee, S.G, Han, S, Jung, J, Jeong, H.S, Hyun, J.K, Rhee, D.K, Kim, H.M, Lee, S. | | Deposit date: | 2020-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.33 Å) | | Cite: | ClpL is a functionally active tetradecameric AAA+ chaperone, distinct from hexameric/dodecameric ones.
Faseb J., 34, 2020
|
|
5YSZ
 
 | | transcriptional regulator CelR-cellobiose complex | | Descriptor: | GLYCEROL, Transcriptional regulator, LacI family, ... | | Authors: | Fu, Y, Yeom, S.Y, Lee, D.H, Lee, S.G. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Structural and functional analyses of the cellulase transcription regulator CelR
FEBS Lett., 592, 2018
|
|
4RCA
 
 | | Crystal structure of human PTPdelta and human Slitrk1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta, SLIT and NTRK-like protein 1, ... | | Authors: | Kim, H.M, Park, B.S, Kim, D, Lee, S.G. | | Deposit date: | 2014-09-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9908 Å) | | Cite: | Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion.
Nat Commun, 5, 2014
|
|
4MAF
 
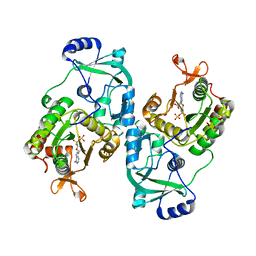 | | Soybean ATP Sulfurylase | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase | | Authors: | Herrmann, J, Ravilious, G.E, McKinney, S.E, Westfall, C.S, Lee, S.G, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and mechanism of soybean ATP sulfurylase and the committed step in plant sulfur assimilation.
J.Biol.Chem., 289, 2014
|
|
4NJ6
 
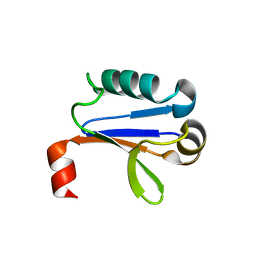 | | PB1 Domain of AtARF7 | | Descriptor: | Auxin response factor 7 | | Authors: | Korasick, D.A, Westfall, C.S, Lee, S.G, Nanao, M, Jez, J.M, Strader, L.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NJ7
 
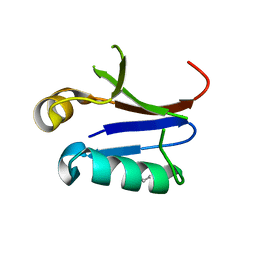 | | PB1 Domain of AtARF7 - SeMet Derivative | | Descriptor: | Auxin response factor 7 | | Authors: | Korasick, D.A, Westfall, C.S, Lee, S.G, Nanao, M, Jez, J.M, Strader, L.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-03-26 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5T9E
 
 | |
5TVS
 
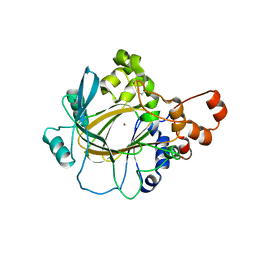 | | JMJD2A in complex with Ni(II) | | Descriptor: | Lysine-specific demethylase 4A, NICKEL (II) ION, ZINC ION | | Authors: | Cascella, B, Lee, S.G, Jez, J.M. | | Deposit date: | 2016-11-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | The small molecule JIB-04 disrupts O2 binding in the Fe-dependent histone demethylase KDM4A/JMJD2A.
Chem. Commun. (Camb.), 53, 2017
|
|
5TVR
 
 | | JMJD2A in complex with Ni(II) and alpha-Ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Cascella, B, Lee, S.G, Jez, J.M. | | Deposit date: | 2016-11-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | The small molecule JIB-04 disrupts O2 binding in the Fe-dependent histone demethylase KDM4A/JMJD2A.
Chem. Commun. (Camb.), 53, 2017
|
|
2HQA
 
 | | Crystal structure of the catalytic alpha subunit of E. Coli replicative DNA polymerase III | | Descriptor: | DNA polymerase III alpha subunit, PHOSPHATE ION | | Authors: | Lamers, M.H, Georgescu, R.E, Lee, S.G, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Cell(Cambridge,Mass.), 126, 2006
|
|
8HGU
 
 | |
8HM5
 
 | |
7XJT
 
 | |
7X99
 
 | |
