5IP5
 
 | |
6N8V
 
 | | Hsp104DWB open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
2KMK
 
 | | Gfi-1 Zinc Fingers 3-5 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*AP*AP*TP*CP*AP*CP*TP*GP*CP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*AP*GP*TP*GP*AP*TP*TP*TP*AP*TP*G)-3'), ZINC ION, ... | | Authors: | Lee, S, Wu, Z. | | Deposit date: | 2009-07-30 | | Release date: | 2010-03-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Gfi-1 zinc domain bound to consensus DNA.
J.Mol.Biol., 397, 2010
|
|
6N8Z
 
 | | HSP104DWB extended conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2019-01-16 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
5VAN
 
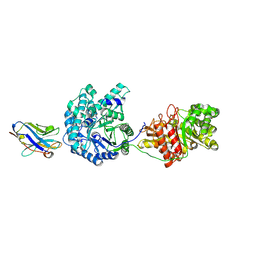 | | Crystal Structure of Beta-Klotho | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
5VAK
 
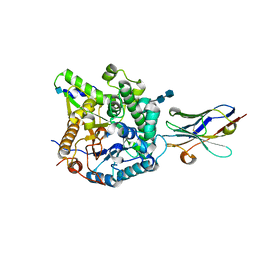 | | Crystal Structure of Beta-Klotho, Domain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-klotho, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
3CRC
 
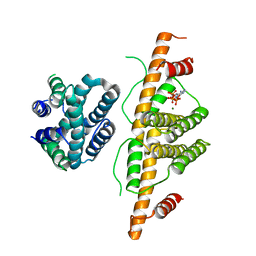 | | Crystal Structure of Escherichia coli MazG, the Regulator of Nutritional Stress Response | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein mazG | | Authors: | Lee, S, Kim, M.H, Kang, B.S, Kim, J.S, Kim, Y.G, Kim, K.J. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response.
J.Biol.Chem., 283, 2008
|
|
3CRA
 
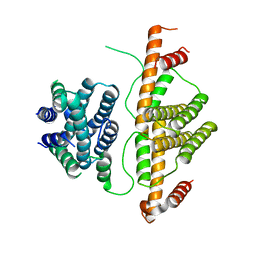 | | Crystal Structure of Escherichia coli MazG, the Regulator of Nutritional Stress Response | | Descriptor: | Protein mazG | | Authors: | Lee, S, Kim, M.H, Kang, B.S, Kim, J.S, Kim, Y.G, Kim, K.J. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response.
J.Biol.Chem., 283, 2008
|
|
5VAQ
 
 | | Crystal Structure of Beta-Klotho in Complex with FGF21CT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-klotho, Fibroblast growth factor 21, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
3EVK
 
 | |
5XD8
 
 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily
Biochem. Biophys. Res. Commun., 491, 2017
|
|
3PMR
 
 | | Crystal Structure of E2 domain of Human Amyloid Precursor-Like Protein 1 | | Descriptor: | Amyloid-like protein 1, PHOSPHATE ION | | Authors: | Lee, S, Xue, Y, Hu, J, Wang, Y, Liu, X, Demeler, B, Ha, Y. | | Deposit date: | 2010-11-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
5CUS
 
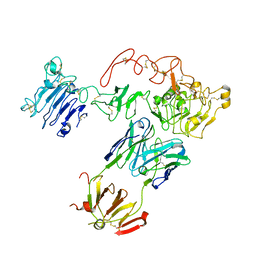 | | Crystal Structure of sErbB3-Fab3379 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab LC region of KTN3379, IgG H chain, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2015-07-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of ErbB3 by a monoclonal antibody that locks the extracellular domain in an inactive configuration.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2OJQ
 
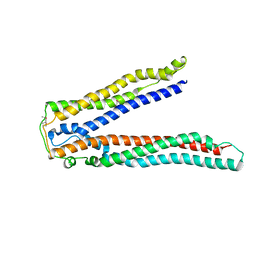 | | Crystal structure of Alix V domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Lee, S, Hurley, J.H. | | Deposit date: | 2007-01-13 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural basis for viral late-domain binding to Alix
Nat.Struct.Mol.Biol., 14, 2007
|
|
5XD7
 
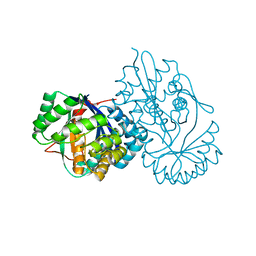 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, ACETIC ACID, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily.
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5XD9
 
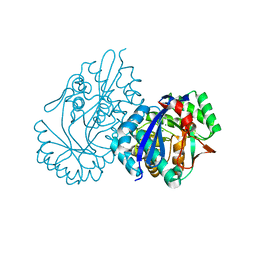 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily
Biochem. Biophys. Res. Commun., 491, 2017
|
|
2FID
 
 | |
2FIF
 
 | |
6MEV
 
 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
6NZ5
 
 | | YcjX-GDPCP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, YcjX Stress Protein | | Authors: | Lee, S, Tsai, J, Tsai, F.T, Sung, N, Lee, J, Chang, C. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
6NZ4
 
 | | YcjX-GDP (type I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, YcjX Stress Protein | | Authors: | Lee, S, Tsai, J, Tsai, F.T. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
5ZCH
 
 | | Crystal structure of OsPP2C50 I267W:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-17 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.474 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
5ZCG
 
 | | Crystal structure of OsPP2C50 S265L/I267V:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
5ZCL
 
 | | Crystal structure of OsPP2C50 I267L:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
5WBW
 
 | | Yeast Hsp104 fragment 1-360 | | Descriptor: | Heat shock protein 104 | | Authors: | Lee, S. | | Deposit date: | 2017-06-29 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural determinants for protein unfolding and translocation by the Hsp104 protein disaggregase.
Biosci. Rep., 37, 2017
|
|
