6KBV
 
 | |
6K8S
 
 | |
6K8U
 
 | |
6K8V
 
 | |
6K8T
 
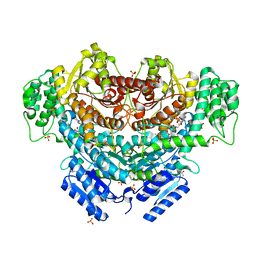 | |
6KBO
 
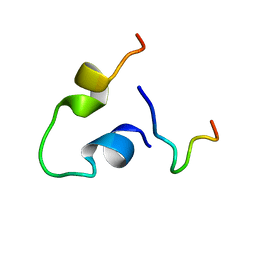 | | Three-dimensional LPS bound structure of VG16KRKP-KYE28. | | Descriptor: | Heparin cofactor 2, VG16KRKP | | Authors: | Ilyas, H, Bhunia, A. | | Deposit date: | 2019-06-26 | | Release date: | 2019-08-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the combinatorial effects of antimicrobial peptides reveal a role of aromatic-aromatic interactions in antibacterial synergism.
J.Biol.Chem., 294, 2019
|
|
2KDU
 
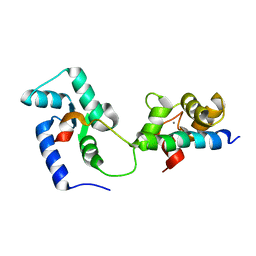 | | Structural basis of the Munc13-1/Ca2+-Calmodulin interaction: A novel 1-26 calmodulin binding motif with a bipartite binding mode | | Descriptor: | CALCIUM ION, Calmodulin, Protein unc-13 homolog A | | Authors: | Rodriguez-Castaneda, F.A, Maestre-Martinez, M, Coudevylle, N, Dimova, K, Jahn, O, Junge, H, Becker, S, Brose, N, Carlomagno, T, Griesinger, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-12-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity.
Embo J., 29, 2010
|
|
4KYZ
 
 | | Three-dimensional structure of triclinic form of de novo design insertion domain, Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | Designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
4KY3
 
 | | Three-dimensional Structure of the orthorhombic crystal of computationally designed insertion domain , Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (2.964 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
4RHP
 
 | | Crystal structure of human COQ9 in complex with a phospholipid, Northeast Structural Genomics Consortium Target HR5043 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Ubiquinone biosynthesis protein COQ9, mitochondrial | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, H, Lee, D, Kogan, S, Maglaqui, M, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-22 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2KLM
 
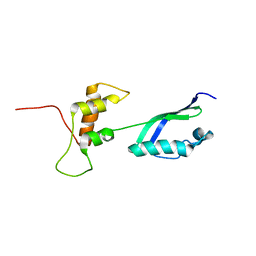 | | Solution Structure of L11 with SAXS and RDC | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Wang, J, Zuo, X, Yu, P, Schwieters, C.D, Wang, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-06 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Determination of multicomponent protein structures in solution using global orientation and shape restraints.
J.Am.Chem.Soc., 131, 2009
|
|
5JXD
 
 | | Crystal structure of murine Tnfaip8 C165S mutant | | Descriptor: | Tumor necrosis factor alpha-induced protein 8, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Park, J, Kim, M.S, Lee, D, Shin, D.H. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The Tnfaip8-PE complex is a novel upstream effector in the anti-autophagic action of insulin
Sci Rep, 7, 2017
|
|
6OE2
 
 | |
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
7ETX
 
 | |
7ETY
 
 | |
7FB6
 
 | | C57D/C146D mutant of Human Cu, Zn Superoxide Dismutase (SOD1) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Baek, Y, Ha, N.-C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the overoxidized Cu/Zn-superoxide dismutase in ROS-induced ALS filament formation.
Commun Biol, 5, 2022
|
|
7FB9
 
 | | Crystal Structure of Human Cu, Zn Superoxide Dismutase (SOD1) | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Baek, Y, Ha, N.-C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of the overoxidized Cu/Zn-superoxide dismutase in ROS-induced ALS filament formation.
Commun Biol, 5, 2022
|
|
3GGL
 
 | | X-Ray Structure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative chitobiase, ZINC ION | | Authors: | Kuzin, A, Neely, H, Seetharaman, R, Lee, D, Ciccosanti, C, Foote, E.L, Janjua, H, Xiao, R, Nair, R, Rost, B, Acton, T, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-28 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Northeast Structural Genomics Consortium Target BtR324A
To be Published
|
|
2KJK
 
 | | Solution structure of the second domain of the listeria protein Lin2157, Northeast Structural Genomics Consortium target Lkr136b | | Descriptor: | Lin2157 protein | | Authors: | Wang, X, Hamilton, K, Xiao, R.H, Lee, D, Ciccosanti, C.H, Nair, R, Rost, B, Acton, T.B, Swapna, G, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Lkr136b
To be Published
|
|
