3Q46
 
 | | Magnesium activated Inorganic pyrophosphatase from Thermococcus thioreducens bound to hydrolyzed product at 0.99 Angstrom resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hughes, R.C, Coates, L, Meehan, E.J, Ng, J.D. | | Deposit date: | 2010-12-23 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Inorganic pyrophosphatase crystals from Thermococcus thioreducens for X-ray and neutron diffraction.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3Q3L
 
 | | The neutron crystallographic structure of inorganic pyrophosphatase from Thermococcus thioreducens | | Descriptor: | CALCIUM ION, Tt-IPPase | | Authors: | Hughes, R.C, Coates, L, Blakeley, M.P, Tomanicek, S.J, Meehan, E.J, Garcia-Ruiz, J.M, Ng, J.D. | | Deposit date: | 2010-12-22 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (2.5 Å) | | Cite: | Inorganic pyrophosphatase crystals from Thermococcus thioreducens for X-ray and neutron diffraction.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3Q4W
 
 | | The structure of archaeal inorganic pyrophosphatase in complex with substrate | | Descriptor: | BROMIDE ION, CALCIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Hughes, R.C, Meehan, E.J, Coates, L, Ng, J.D. | | Deposit date: | 2010-12-24 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Inorganic pyrophosphatase crystals from Thermococcus thioreducens for X-ray and neutron diffraction.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Kovalevsky, A.Y, Gerlits, O.O. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6K9R
 
 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
5E5K
 
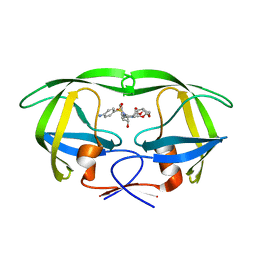 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Kovalevsky, A.Y, Das, A. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3TMJ
 
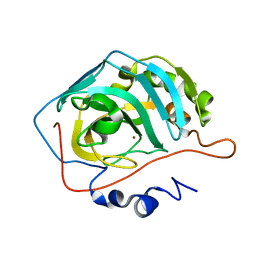 | | Joint X-ray/neutron structure of human carbonic anhydrase II at pH 7.8 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Fisher, Z. | | Deposit date: | 2011-08-31 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Neutron Structure of Human Carbonic Anhydrase II: A Hydrogen-Bonded Water Network "Switch" Is Observed between pH 7.8 and 10.0.
Biochemistry, 50, 2011
|
|
3U7T
 
 | |
1LZN
 
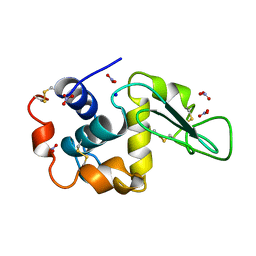 | | NEUTRON STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | NITRATE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Bon, C.I, Lehmann, M.S, Wilkinson, C. | | Deposit date: | 1999-03-23 | | Release date: | 1999-04-01 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å) | | Cite: | Quasi-Laue neutron-diffraction study of the water arrangement in crystals of triclinic hen egg-white lysozyme.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7D49
 
 | |
7D3Z
 
 | |
7D4L
 
 | |
7D4X
 
 | |
7D6G
 
 | |
7JY2
 
 | | Z-DNA joint X-ray/Neutron | | Descriptor: | Chains: A,B | | Authors: | Harp, J.M, Coates, L, Egli, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
7LB7
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A.Y, Kneller, D.W, Coates, L. | | Deposit date: | 2021-01-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography.
J.Med.Chem., 64, 2021
|
|
3KMF
 
 | |
4PSY
 
 | | 100K crystal structure of Escherichia coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Wilson, M.A, Wan, Q, Bennet, B.C, Dealwis, C, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 22, 2014
|
|
4RGC
 
 | | 277K Crystal structure of Escherichia Coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Wilson, M.A, Wan, Q, Bennett, B.C, Dealwis, C. | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5T8H
 
 | |
5WEY
 
 | | Joint X-ray/neutron structure of Concanavalin A with alpha1-2 D-mannobiose | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Woods, R.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | Mannobiose Binding Induces Changes in Hydrogen Bonding and Protonation States of Acidic Residues in Concanavalin A As Revealed by Neutron Crystallography.
Biochemistry, 56, 2017
|
|
4FC1
 
 | |
4IAK
 
 | |
4IAY
 
 | |
4IAF
 
 | |
