2QQU
 
 | | Crystal structure of a cell-wall invertase (D239A) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, ... | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Rabijns, A, Van den Ende, W. | | Deposit date: | 2007-07-27 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of Arabidopsis thaliana cell-wall invertase mutants in complex with sucrose.
J.Mol.Biol., 377, 2008
|
|
2QQW
 
 | | Crystal structure of a cell-wall invertase (D23A) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, ... | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Rabijns, A, Van den Ende, W. | | Deposit date: | 2007-07-27 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Arabidopsis thaliana cell-wall invertase mutants in complex with sucrose.
J.Mol.Biol., 377, 2008
|
|
3UGF
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
3UGH
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 6-kestose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
3UGG
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 1-kestose | | Descriptor: | GLYCEROL, SULFATE ION, Sucrose:(Sucrose/fructan) 6-fructosyltransferase, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
2QQV
 
 | | Crystal structure of a cell-wall invertase (E203A) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, ... | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Rabijns, A, Van den Ende, W. | | Deposit date: | 2007-07-27 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of Arabidopsis thaliana cell-wall invertase mutants in complex with sucrose.
J.Mol.Biol., 377, 2008
|
|
2OXB
 
 | | Crystal structure of a cell-wall invertase (E203Q) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2007-02-20 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alternate sucrose binding mode in the E203Q Arabidopsis invertase mutant: An X-ray crystallography and docking study.
Proteins, 71, 2007
|
|
2AEZ
 
 | | Crystal structure of fructan 1-exohydrolase IIa (E201Q) from Cichorium intybus in complex with 1-kestose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Verhaest, M, Lammens, W, Le Roy, K, De Ranter, C.J, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor.
New Phytol, 174, 2007
|
|
2AQU
 
 | | Structure of HIV-1 protease bound to atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, HIV-1 Protease | | Authors: | Clemente, J.C, Coman, R.M, Thiaville, M.M, Janka, L.K, Jeung, J.A, Nukoolkarn, S, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Leelamanit, W, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2005-08-18 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of HIV-1 CRF_01 A/E protease inhibitor resistance: structural determinants for maintaining sensitivity and developing resistance to atazanavir.
Biochemistry, 45, 2006
|
|
2ERV
 
 | | Crystal structure of the outer membrane enzyme PagL | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL MONODECYL ETHER, hypothetical protein Paer03002360 | | Authors: | Rutten, L, Geurtsen, J, Lambert, W, Smolenaers, J.J, Bonvin, A.M, van der Ley, P, Egmond, M.R, Gros, P, Tommassen, J. | | Deposit date: | 2005-10-25 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3G7M
 
 | | Structure of the thaumatin-like xylanase inhibitor TLXI | | Descriptor: | GLYCEROL, SODIUM ION, Xylanase inhibitor TL-XI | | Authors: | Vandermarliere, E, Courtin, C.M, Lammens, W, Schoepe, J, Strelkov, S.V. | | Deposit date: | 2009-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of the noncompetitive xylanase inhibitor TLXI, member of the small thaumatin-like protein family.
Proteins, 78, 2010
|
|
2XQR
 
 | | Crystal structure of plant cell wall invertase in complex with a specific protein inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hothorn, M, Van den Ende, W, Lammens, W, Rybin, V, Scheffzek, K. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural Insights Into the Ph-Controlled Targeting of Plant Cell-Wall Invertase by a Specific Inhibitor Protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4PNJ
 
 | | Recombinant Sperm Whale P6 Myoglobin Solved with Single Pulse Free Electron Laser Data | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Cohen, A, Gonzalez, A, Lam, W, Lyubimov, A, Sauter, N, Tsai, Y, Uervirojnangkoorn, M, Brunger, A, Soltis, M. | | Deposit date: | 2014-05-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Goniometer-based femtosecond crystallography with X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6QO8
 
 | |
6QO9
 
 | | Crystal structure of ribonucleotide reductase NrdF from Bacillus anthracis soaked with manganese ions | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, Ribonucleoside-diphosphate reductase subunit beta, ... | | Authors: | Grave, K, Hogbom, M. | | Deposit date: | 2019-02-12 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Redox-induced structural changes in the di-iron and di-manganese forms of Bacillus anthracis ribonucleotide reductase subunit NrdF suggest a mechanism for gating of radical access.
J.Biol.Inorg.Chem., 24, 2019
|
|
6QO5
 
 | |
6QOB
 
 | |
6QO7
 
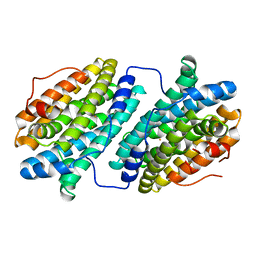 | |
2ADE
 
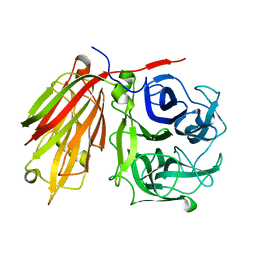 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus in complex with fructose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-fructofuranose, ... | | Authors: | Verhaest, M, Le Roy, K, De Ranter, C.J, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor.
New Phytol, 174, 2007
|
|
2ADD
 
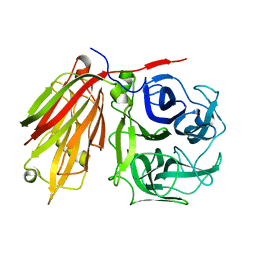 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Verhaest, M, Le Roy, K, De Ranter, C.J, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor.
New Phytol, 174, 2007
|
|
2AC1
 
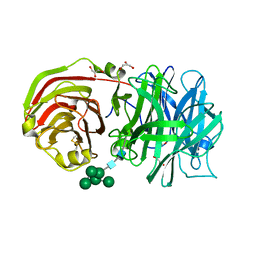 | | Crystal structure of a cell-wall invertase from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Verhaest, M, Le Roy, K, De Ranter, C, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-18 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray diffraction structure of a cell-wall invertase from Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2AEY
 
 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus in complex with 2,5 dideoxy-2,5-immino-D-mannitol | | Descriptor: | 2,5-DIDEOXY-2,5-IMINO-D-MANNITOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Verhaest, M, Le Roy, K, De Ranter, C.J, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor.
New Phytol, 174, 2007
|
|
6TTJ
 
 | | Neutral invertase 2 from Arabidopsis thaliana | | Descriptor: | Alkaline/neutral invertase CINV1 | | Authors: | Tsirkone, V.G, Osipov, E.M, Beelen, S, Strelkov, S.V. | | Deposit date: | 2019-12-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.392 Å) | | Cite: | Crystal structure of Arabidopsis thaliana neutral invertase 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1GNO
 
 | | HIV-1 PROTEASE (WILD TYPE) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
1GNN
 
 | | HIV-1 PROTEASE MUTANT WITH VAL 82 REPLACED BY ASN (V82N) COMPLEXED WITH U89360E (INHIBITOR) | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Treharne, A, Hartsuck, J.A, Foundling, S, Tang, J. | | Deposit date: | 1996-05-04 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases.
Biochemistry, 35, 1996
|
|
