1Y1U
 
 | | Structure of unphosphorylated STAT5a | | Descriptor: | Signal transducer and activator of transcription 5A | | Authors: | Neculai, D, Neculai, A.M, Verrier, S, Straub, K, Klumpp, K, Pfitzner, E, Becker, S. | | Deposit date: | 2004-11-19 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structure of the unphosphorylated STAT5a dimer
J.Biol.Chem., 280, 2005
|
|
4F7B
 
 | | Structure of the lysosomal domain of limp-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Neculai, D, Ravichandran, M, Seitova, A, Neculai, M, Pizzaro, J.C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-15 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36.
Nature, 504, 2013
|
|
3IV1
 
 | | Coiled-coil domain of tumor susceptibility gene 101 | | Descriptor: | CHLORIDE ION, SULFATE ION, Tumor susceptibility gene 101 protein | | Authors: | Neculai, D, Avvakumov, G.V, Wernimont, A.K, Xue, S, Walker, J.R, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coiled-Coil Domain of Human Tsg101
To be Published
|
|
5BNH
 
 | | Crystal structure of the HLTF HIRAN domain with a ssDNA fragment | | Descriptor: | ACETATE ION, DNA (5'-D(*(GD)P*GP*TP*G)-3'), DNA (5'-D(*(TD)P*TP*G)-3'), ... | | Authors: | Neculai, D, Walker, J.R, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structure of the HLTF HIRAN domain with a ssDNA fragment
To Be Published
|
|
2QGX
 
 | | Ubiquitin-conjugating enzyme E2Q | | Descriptor: | Ubiquitin-conjugating enzyme E2 Q1 | | Authors: | Neculai, D, Avvakumov, G.V, Xue, S, Walker, J.R, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Sicheri, F, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
7R3K
 
 | | Chlamydomonas reinhardtii TSP9 mutant small Photosystem I complex | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R)-beta,beta-caroten-3-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Klaiman, D, Schwartz, T, Nelson, N. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structure of Photosystem I Supercomplex Isolated from a Chlamydomonas reinhardtii Cytochrome b6f Temperature-Sensitive Mutant.
Biomolecules, 13, 2023
|
|
3I3T
 
 | | Crystal structure of covalent ubiquitin-USP21 complex | | Descriptor: | ETHANAMINE, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 21, ... | | Authors: | Neculai, D, Avvakumov, G.V, Walker, J.R, Xue, S, Butler-Cole, C, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3ENO
 
 | |
3ENH
 
 | | Crystal structure of Cgi121/Bud32/Kae1 complex | | Descriptor: | HEXATANTALUM DODECABROMIDE, Putative O-sialoglycoprotein endopeptidase, Uncharacterized protein MJ0187 | | Authors: | Neculai, D. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Atomic Structure of the KEOPS Complex: An Ancient Protein Kinase-Containing Molecular Machine
Mol.Cell, 32, 2008
|
|
3ENC
 
 | | Crystal structure of Pyrococcus furiosus PCC1 dimer | | Descriptor: | protein PCC1 | | Authors: | Neculai, D. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6304 Å) | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
3EN9
 
 | | Structure of the Methanococcus jannaschii KAE1-BUD32 fusion protein | | Descriptor: | HEXATANTALUM DODECABROMIDE, MAGNESIUM ION, O-sialoglycoprotein endopeptidase/protein kinase | | Authors: | Neculai, D. | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
4K25
 
 | | Crystal Structure of yeast Qri7 homodimer | | Descriptor: | CALCIUM ION, Probable tRNA threonylcarbamoyladenosine biosynthesis protein QRI7, mitochondrial, ... | | Authors: | Neculai, D, Wan, L, Mao, D.Y, Sicheri, F. | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-08 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system.
Nucleic Acids Res., 41, 2013
|
|
6QPH
 
 | | Dunaliella minimal PSI complex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Klaiman, D, Caspy, I, Nelson, N. | | Deposit date: | 2019-02-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a minimal photosystem I from the green alga Dunaliella salina.
Nat.Plants, 6, 2020
|
|
2P64
 
 | | D domain of b-TrCP | | Descriptor: | CADMIUM ION, F-box/WD repeat protein 1A | | Authors: | Neculai, D, Orlicky, S, Ceccarelli, D. | | Deposit date: | 2007-03-16 | | Release date: | 2007-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination.
Cell(Cambridge,Mass.), 129, 2007
|
|
3L11
 
 | | Crystal Structure of the Ring Domain of RNF168 | | Descriptor: | E3 ubiquitin-protein ligase RNF168, MALONATE ION, ZINC ION | | Authors: | Neculai, D, Yermekbayeva, L, Crombet, L, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-10 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation.
J.Biol.Chem., 287, 2012
|
|
3PDF
 
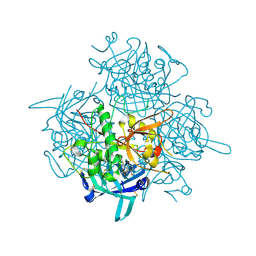 | | Discovery of Novel Cyanamide-Based Inhibitors of Cathepsin C | | Descriptor: | 2,5-dibromo-N-{(3R,5S)-1-[(Z)-iminomethyl]-5-methylpyrrolidin-3-yl}benzenesulfonamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, B, Laine, D. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of novel cyanamide-based inhibitors of cathepsin C.
Acs Med.Chem.Lett., 2, 2011
|
|
2OB4
 
 | | Human Ubiquitin-Conjugating Enzyme CDC34 | | Descriptor: | Ubiquitin-conjugating enzyme E2-32 kDa complementing | | Authors: | Neculai, D, Avvakumov, G.V, Xue, S, Walker, J.R, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Sicheri, F, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-18 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
5K5F
 
 | | NMR structure of the HLTF HIRAN domain | | Descriptor: | Helicase-like transcription factor | | Authors: | Bezsonova, I, Neculai, D, Korzhnev, D, Weigelt, J, Bountra, C, Edwards, A, Arrowsmith, C, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HLTF HIRAN domain
To Be Published
|
|
1XA8
 
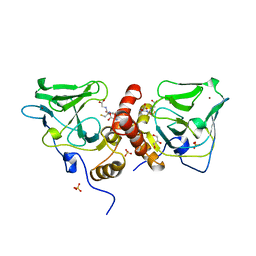 | | Crystal Structure Analysis of Glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, ... | | Authors: | Neculai, A.M, Neculai, D, Griesinger, C, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
1X6M
 
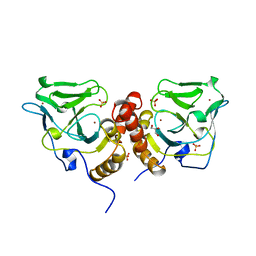 | | Crystal structure of the glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, SULFATE ION, ... | | Authors: | Neculai, A.M, Neculai, D, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
3ENP
 
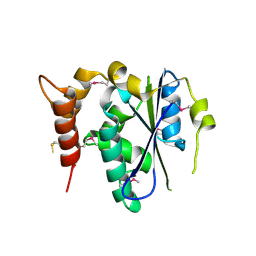 | | Crystal structure of human cgi121 | | Descriptor: | TP53RK-binding protein | | Authors: | Haffani, Y.Z, Ceccarelli, D.F, Neculai, D, Mao, D.Y, Sicheri, F. | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
4CIS
 
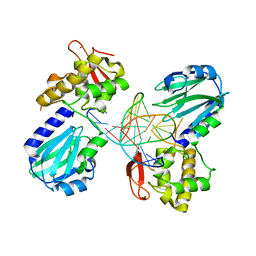 | | Structure of MutM in complex with carbocyclic 8-oxo-G containing DNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, DNA, FORMAMIDOPYRIMIDIN DNA GLYCOSYLASE, ... | | Authors: | Schneider, S, Sadeghian, K, Flaig, D, Blank, I.D, Strasser, R, Stathis, D, Winnacker, M, Carell, T, Ochsenfeld, C. | | Deposit date: | 2013-12-15 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ribose-protonated DNA base excision repair: a combined theoretical and experimental study.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
6RHZ
 
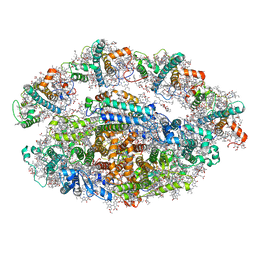 | | Structure of a minimal photosystem I from a green alga | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Perez Boerema, A, Klaiman, D, Caspy, I, Netzer-El, S.Y, Amunts, A, Nelson, N. | | Deposit date: | 2019-04-23 | | Release date: | 2020-02-19 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a minimal photosystem I from the green alga Dunaliella salina.
Nat.Plants, 6, 2020
|
|
2K8Y
 
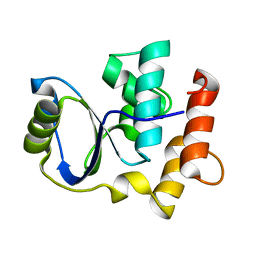 | | Solution NMR Structure of Cgi121 from Methanococcus jannaschii. Northeast Structural Genomics Consortium Target MJ0187 | | Descriptor: | Uncharacterized protein MJ0187 | | Authors: | Rumpel, S, Fares, C, Neculai, D, Arrowsmith, C, Montelione, G.T, Sicheri, F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
7BLZ
 
 | | Red alga C.merolae Photosystem I | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R)-beta,beta-caroten-3-ol, ... | | Authors: | Nelson, N, Klaiman, D, Hippler, M. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Red alga C.merolae Photosystem I
To Be Published
|
|
