1R0S
 
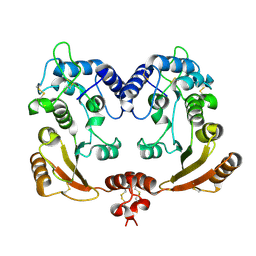 | | Crystal structure of ADP-ribosyl cyclase Glu179Ala mutant | | Descriptor: | ADP-ribosyl cyclase | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-22 | | Release date: | 2004-03-09 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
1R16
 
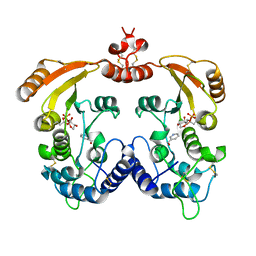 | | Aplysia ADP ribosyl cyclase with bound pyridylcarbinol and R5P | | Descriptor: | 3-PYRIDINYLCARBINOL, ADP-ribosyl cyclase, ANY 5'-MONOPHOSPHATE NUCLEOTIDE | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-23 | | Release date: | 2004-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
1R15
 
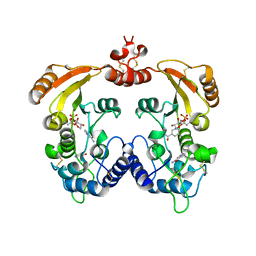 | | Aplysia ADP ribosyl cyclase with bound nicotinamide and R5P | | Descriptor: | ADP-ribosyl cyclase, ANY 5'-MONOPHOSPHATE NUCLEOTIDE, NICOTINAMIDE | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-23 | | Release date: | 2004-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
3F6Y
 
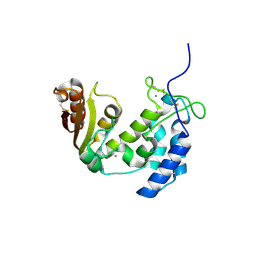 | | Conformational Closure of the Catalytic Site of Human CD38 Induced by Calcium | | Descriptor: | ADP-ribosyl cyclase 1, CALCIUM ION | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Lam, C.M.C, Lee, H.C, Hao, Q. | | Deposit date: | 2008-11-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational Closure of the Catalytic Site of Human CD38 Induced by Calcium
Biochemistry, 47, 2008
|
|
2HCT
 
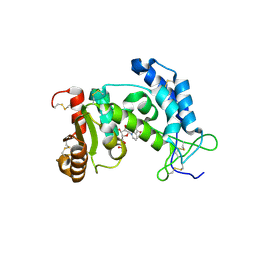 | | Acidic residues at the active sites of CD38 and ADP-ribosyl cyclase determine NAAPD synthesis and hydrolysis activities | | Descriptor: | ADP-ribosyl cyclase 1, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE | | Authors: | Liu, Q, Kriksunov, I.A, Hao, Q, Graeff, R, Lee, H.C. | | Deposit date: | 2006-06-18 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Acidic residues at the active sites of CD38 and ADP-ribosyl cyclase determine nicotinic acid adenine dinucleotide phosphate (NAADP) synthesis and hydrolysis activities.
J.Biol.Chem., 281, 2006
|
|
2I67
 
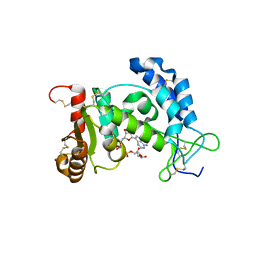 | | Structural Basis for the Mechanistic Understanding Human CD38 Controlled Multiple Catalysis | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribosyl cyclase 1 | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis.
J.Biol.Chem., 281, 2006
|
|
2I66
 
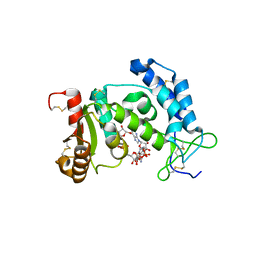 | | Structural Basis for the Mechanistic Understanding Human CD38 Controlled Multiple Catalysis | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3R,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4R,5S)-3,4,5-TRIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, [(2R,3R,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis.
J.Biol.Chem., 281, 2006
|
|
2I65
 
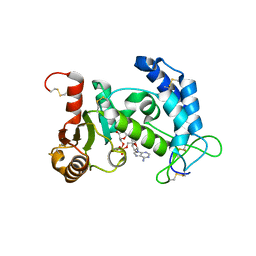 | | Structural Basis for the Mechanistic Understanding Human CD38 Controlled Multiple Catalysis | | Descriptor: | ADP-ribosyl cyclase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis.
J.Biol.Chem., 281, 2006
|
|
2FRH
 
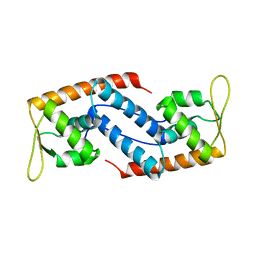 | | Crystal Structure of Sara, A Transcription Regulator From Staphylococcus Aureus | | Descriptor: | CALCIUM ION, Staphylococcal accessory regulator A | | Authors: | Liu, Y, Manna, A.C, Ingavale, S, Cheung, A.L, Zhang, G. | | Deposit date: | 2006-01-19 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FNP
 
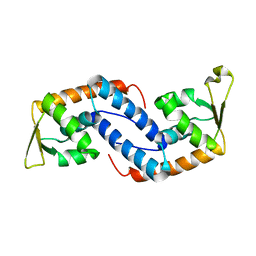 | | Crystal structure of SarA | | Descriptor: | Staphylococcal accessory regulator A | | Authors: | Liu, Y, Manna, A.C, Pan, C.H, Cheung, A.L, Zhang, G. | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
