5TMU
 
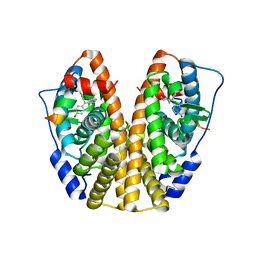 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-(cycloheptylidenemethylene)diphenol | | Descriptor: | 4,4'-(cycloheptylidenemethylene)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN4
 
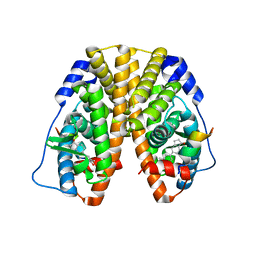 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the ACD-ring estrogen, (S)-5-(4-hydroxy-3,5-dimethylphenyl)-2,3-dihydro-1H-inden-1-ol | | Descriptor: | (1S)-5-(4-hydroxy-3,5-dimethylphenyl)-2,3-dihydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM9
 
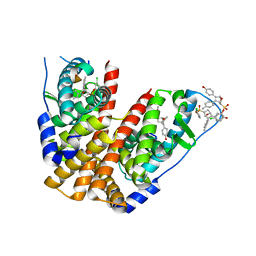 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, (E)-3-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)acrylic acid | | Descriptor: | 3-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}prop-2-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM1
 
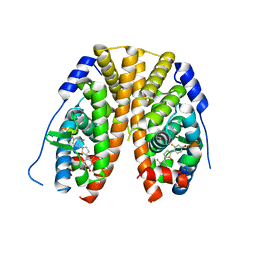 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,5-bis(2-fluoro-4-hydroxyphenyl)thiophene 1-oxide | | Descriptor: | 2,5-bis(2-fluoro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMR
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Cyclofenil-ASC derivative, ethyl (E)-3-(4-(cyclohexylidene(4-hydroxyphenyl)methyl)phenyl)acrylate | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, ethyl 3-{4-[cyclohexylidene(4-hydroxyphenyl)methyl]phenyl}prop-2-enoate | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the estradiol derivative, (8S,9S,13S,14S,E)-17-((4-isopropylphenyl)imino)-13-methyl-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthren-3-ol | | Descriptor: | (9beta,13alpha,17Z)-17-{[4-(propan-2-yl)phenyl]imino}estra-1,3,5(10)-trien-3-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5UGM
 
 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with Edaglitazone | | Descriptor: | (5R)-5-({4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}methyl)-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPAR gamma.
Elife, 7, 2018
|
|
4R2U
 
 | | Crystal Structure of PPARgamma in complex with SR1664 | | Descriptor: | 4'-[(2,3-dimethyl-5-{[(1S)-1-(4-nitrophenyl)ethyl]carbamoyl}-1H-indol-1-yl)methyl]biphenyl-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Marciano, D.P, Kamenecka, T, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-08-13 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological repression of PPAR gamma promotes osteogenesis.
Nat Commun, 6, 2015
|
|
4R6S
 
 | | Crystal structure of PPARgammma in complex with SR1663 | | Descriptor: | 4'-[(2,3-dimethyl-5-{[(1R)-1-(4-nitrophenyl)ethyl]carbamoyl}-1H-indol-1-yl)methyl]biphenyl-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Marciano, D.P, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-08-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Pharmacological repression of PPAR gamma promotes osteogenesis.
Nat Commun, 6, 2015
|
|
4N73
 
 | |
6N9G
 
 | | Crystal Structure of RGS7-Gbeta5 dimer | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Regulator of G-protein signaling 7 | | Authors: | Patil, D.N, Rangarajan, E, Izard, T, Martemyanov, K.A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Structural organization of a major neuronal G protein regulator, the RGS7-G beta 5-R7BP complex.
Elife, 7, 2018
|
|
2RO3
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator Abh | | Descriptor: | Putative transition state regulator abh | | Authors: | Sullivan, D.M, Bobay, B.G, Douglas, K.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2012-04-04 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
2K1N
 
 | | DNA bound structure of the N-terminal domain of AbrB | | Descriptor: | AbrB family transcriptional regulator, DNA (25-MER) | | Authors: | Cavanagh, J, Bobay, B.G, Sullivan, D.M, Thompson, R.J. | | Deposit date: | 2008-03-10 | | Release date: | 2008-11-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Insights into the Nature of DNA Binding of AbrB-like Transcription Factors
Structure, 16, 2008
|
|
5CC1
 
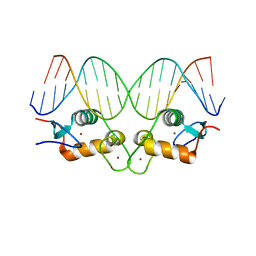 | | S425G Glucocorticoid receptor DNA binding domain - (+)GRE complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Weikum, E.A, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CBX
 
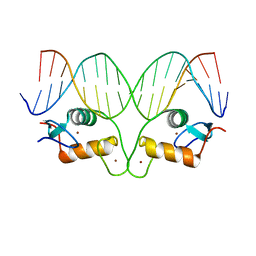 | | AncGR DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncGR DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CBY
 
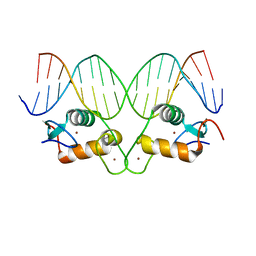 | | AncGR2 DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncGR2 DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CBZ
 
 | | AncMR DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncMR DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CC0
 
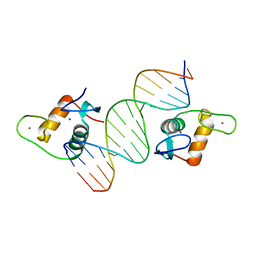 | | AncSR2 - TSLP nGRE complex | | Descriptor: | AncSR2 DNA Binding Domain, DNA (5'-D(*AP*GP*CP*TP*CP*TP*CP*CP*CP*GP*GP*AP*GP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*CP*TP*CP*CP*GP*GP*GP*AP*GP*AP*GP*CP*T)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5E6A
 
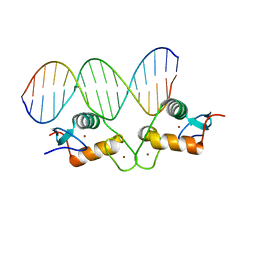 | | Glucocorticoid receptor DNA binding domain - PLAU NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*TP*GP*AP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
5E69
 
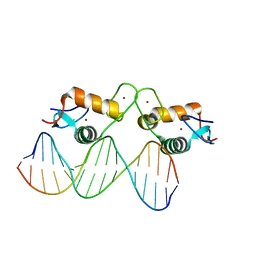 | | Glucocorticoid receptor DNA binding domain - IL8 NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*CP*GP*AP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
5E6D
 
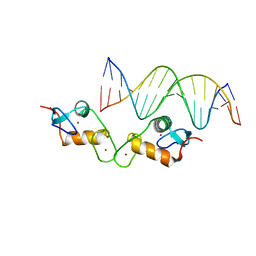 | | Glucocorticoid receptor DNA binding domain - ICAM1 NF-kB response element complex | | Descriptor: | DNA (5'-D(*GP*CP*TP*CP*CP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*GP*GP*AP*GP*C)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
5E6C
 
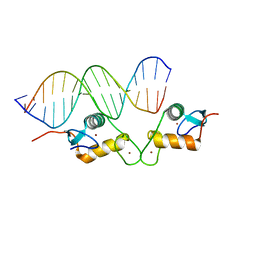 | | Glucocorticoid receptor DNA binding domain - CCL2 NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*GP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
5E6B
 
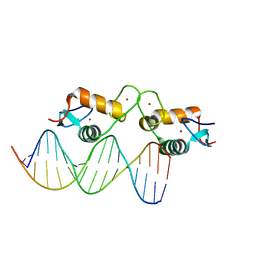 | | Glucocorticoid receptor DNA binding domain - RELB NF-kB response element complex | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*GP*AP*AP*TP*TP*CP*CP*GP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*CP*GP*GP*AP*AP*TP*TP*CP*CP*CP*CP*GP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
4MCE
 
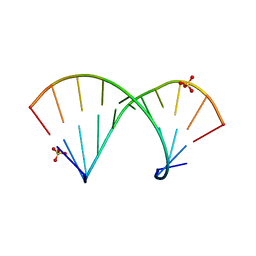 | | Crystal structure of the Gas5 GRE Mimic | | Descriptor: | Gas5 GREM Fwd, Gas5 GREM Rev, SULFATE ION | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2013-08-21 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate.
Nat Commun, 5, 2014
|
|
4MCF
 
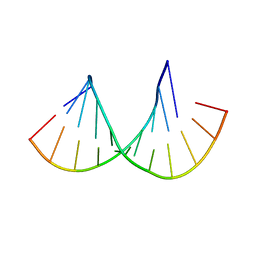 | | Crystal structure of the Gas5 GRE Mimic | | Descriptor: | Gas5 GREM Fwd, Gas5 GREM Rev, SULFATE ION | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2013-08-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate.
Nat Commun, 5, 2014
|
|
