4TRW
 
 | | Structure of BACE1 complex with a syn-HEA-type inhibitor | | Descriptor: | Beta-secretase 1, L-alpha-glutamyl-L-isoleucyl-N-[(2R,3S)-1-{[(1S)-1-carboxybutyl]amino}-2-hydroxy-5-methylhexan-3-yl]-3-thiophen-2-yl-L-alaninamide | | Authors: | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evaluation of transition-state mimics in a superior BACE1 cleavage sequence as peptide-mimetic BACE1 inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TRZ
 
 | | Structure of BACE1 complex with 2-thiophenyl HEA-type inhibitor | | Descriptor: | 2-thiophenyl HEA-type inhibitor, Beta-secretase 1 | | Authors: | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Evaluation of transition-state mimics in a superior BACE1 cleavage sequence as peptide-mimetic BACE1 inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4WY3
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3R,4aS,8aR)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-11-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6R
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6T
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1AO6
 
 | |
1NDH
 
 | |
7DH5
 
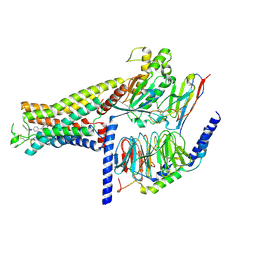 | | Dog beta3 adrenergic receptor bound to mirabegron in complex with a miniGs heterotrimer | | Descriptor: | 2-(2-azanyl-1,3-thiazol-4-yl)-N-[4-[2-[[(2R)-2-oxidanyl-2-phenyl-ethyl]amino]ethyl]phenyl]ethanamide, Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-11-12 | | Release date: | 2021-08-04 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structure of the beta 3-adrenergic receptor reveals the molecular basis of subtype selectivity.
Mol.Cell, 81, 2021
|
|
7XJH
 
 | | Isoproterenol-activated dog beta3 adrenergic receptor | | Descriptor: | Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the beta 3-adrenergic receptor reveals the molecular basis of subtype selectivity.
Mol.Cell, 81, 2021
|
|
6NBJ
 
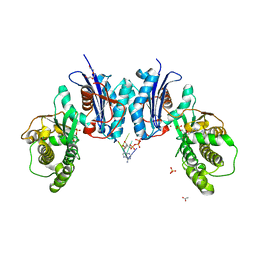 | | Qri7 | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stec, B. | | Deposit date: | 2018-12-07 | | Release date: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Discovery of the Universal tRNA Binding Mode for the TsaD-like Components of the t6A tRNA Modification Pathway
Biophysica, 3, 2023
|
|
5C5O
 
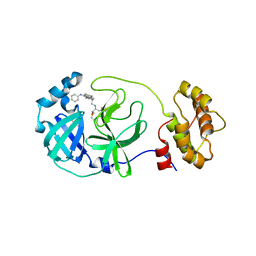 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3S,4aR,8aS)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
5C5N
 
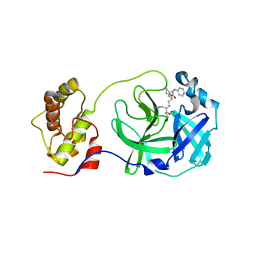 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3R,4aS,8aR)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
1FU3
 
 | |
3RP2
 
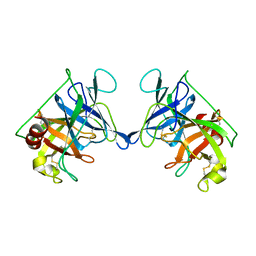 | | THE STRUCTURE OF RAT MAST CELL PROTEASE II AT 1.9-ANGSTROMS RESOLUTION | | Descriptor: | RAT MAST CELL PROTEASE II | | Authors: | Reynolds, R, Remington, S, Weaver, L, Fischer, R, Anderson, W, Ammon, H, Matthews, B. | | Deposit date: | 1984-09-10 | | Release date: | 1984-10-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of rat mast cell protease II at 1.9-A resolution.
Biochemistry, 27, 1988
|
|
5GGF
 
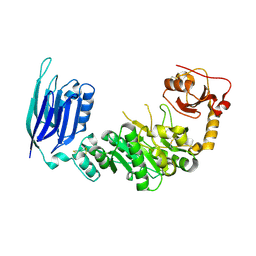 | |
5GGG
 
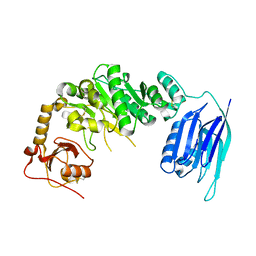 | |
5GGN
 
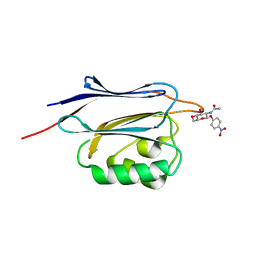 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGJ
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Man-alpha-pNP | | Descriptor: | 4-nitrophenyl alpha-D-mannopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGL
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-alpha-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGP
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta1,2-Man-peptide | | Descriptor: | 10-mer Peptide from Dystroglycan, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGI
 
 | | Crystal structure of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Mn, UDP and Mannosyl-peptide | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGK
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Man-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-mannopyranoside, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGO
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GalNac-beta1,3-GlcNAc-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7CTQ
 
 | | Peptidyl tryptophan dihydroxylase QhpG essential for tryptophylquinone cofactor biogenesis | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-6-(cyclohexylmethoxy)-2-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-3-yl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol, FLAVIN-ADENINE DINUCLEOTIDE, HEXANE-1,6-DIOL, ... | | Authors: | Oozeki, T, Nakai, T, Okajima, T. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Functional and structural characterization of a flavoprotein monooxygenase essential for biogenesis of tryptophylquinone cofactor.
Nat Commun, 12, 2021
|
|
