5A34
 
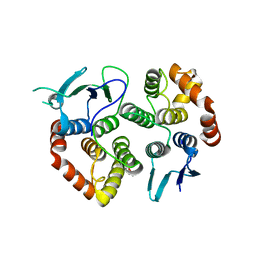 | | The crystal structure of the GST-like domains complex of EPRS-AIMP2 | | Descriptor: | AMINOACYL TRNA SYNTHASE COMPLEX-INTERACTING MULTIFUNCTIONAL PROTEIN 2, BIFUNCTIONAL GLUTAMATE/PROLINE--TRNA LIGASE, GLYCEROL | | Authors: | Cho, H.Y, Kang, B.S. | | Deposit date: | 2015-05-27 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Assembly of Multi-tRNA Synthetase Complex Via Heterotetrameric Glutathione Transferase-Homology Domains.
J.Biol.Chem., 290, 2015
|
|
5BMU
 
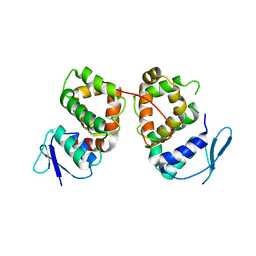 | |
5HWT
 
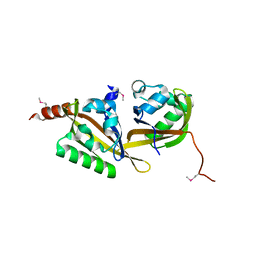 | | Crystal structure of apo-PAS1 | | Descriptor: | Sensor histidine kinase TodS | | Authors: | Hwang, J, Koh, S. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Insights into Toluene Sensing in the TodS/TodT Signal Transduction System.
J. Biol. Chem., 291, 2016
|
|
5HWW
 
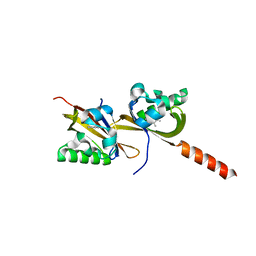 | | Crystal structure of PAS1 complexed with 1,2,4-TMB | | Descriptor: | 1,2,4-trimethylbenzene, Sensor histidine kinase TodS | | Authors: | Hwang, J, Koh, S. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Insights into Toluene Sensing in the TodS/TodT Signal Transduction System.
J. Biol. Chem., 291, 2016
|
|
5HWV
 
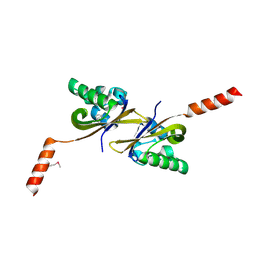 | | Crystal structure of PAS1 complexed with toluene | | Descriptor: | Sensor histidine kinase TodS, TOLUENE | | Authors: | Hwang, J, Koh, S. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular Insights into Toluene Sensing in the TodS/TodT Signal Transduction System.
J. Biol. Chem., 291, 2016
|
|
6JLB
 
 | |
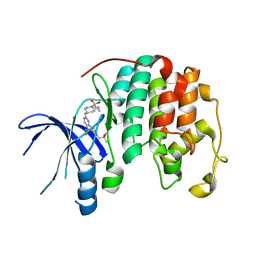
8UV0
 
 | |
1DU3
 
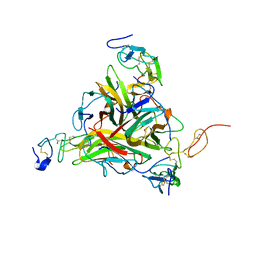 | | Crystal structure of TRAIL-SDR5 | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND, ZINC ION | | Authors: | Cha, S.-S, Sung, B.-J, Oh, B.-H. | | Deposit date: | 2000-01-14 | | Release date: | 2000-09-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity
J.Biol.Chem., 275, 2000
|
|
5JU6
 
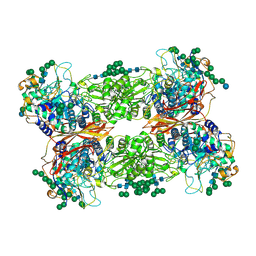 | | Structural and Functional Studies of Glycoside Hydrolase Family 3 beta-Glucosidase Cel3A from the Moderately Thermophilic Fungus Rasamsonia emersonii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Gudmundsson, M, Sandgren, M, Karkehabadi, S. | | Deposit date: | 2016-05-10 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional studies of the glycoside hydrolase family 3 beta-glucosidase Cel3A from the moderately thermophilic fungus Rasamsonia emersonii.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6ICI
 
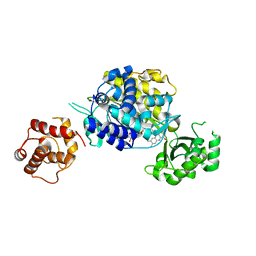 | | Crystal structure of human MICAL3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, [F-actin]-monooxygenase MICAL3 | | Authors: | Hwang, K.Y, Kim, J.S. | | Deposit date: | 2018-09-06 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic insights into flavin-containing monooxygenase and calponin-homology domains in human MICAL3.
Iucrj, 7, 2020
|
|
5H3H
 
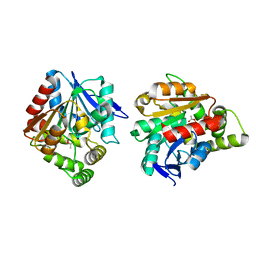 | | Esterase (EaEST) from Exiguobacterium antarcticum | | Descriptor: | Abhydrolase domain-containing protein, ETHANEPEROXOIC ACID | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-10-24 | | Release date: | 2017-01-11 | | Last modified: | 2022-10-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Functional Characterization of an Esterase (EaEST) from Exiguobacterium antarcticum.
Plos One, 12, 2017
|
|
5HZT
 
 | | Crystal structure of Dronpa-Cu2+ | | Descriptor: | COPPER (II) ION, Fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5HZU
 
 | | Crystal structure of Dronpa-Ni2+ | | Descriptor: | Fluorescent protein Dronpa, NICKEL (II) ION | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5HZS
 
 | | Crystal structure of Dronpa-Co2+ | | Descriptor: | COBALT (II) ION, Fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
7EC1
 
 | | Crystal structure of SdgB (ligand-free form) | | Descriptor: | GLYCEROL, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-24 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC3
 
 | | Crystal structure of SdgB (complexed with UDP, GlcNAc, and Glycosylated peptide) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-35)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-65)]5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC6
 
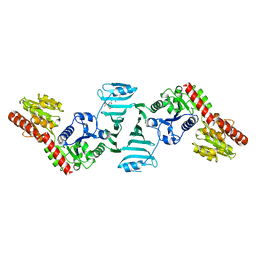 | | Crystal structure of SdgB (complexed with peptides) | | Descriptor: | ASP-SER-ASP, Glycosyl transferase, group 1 family protein | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC7
 
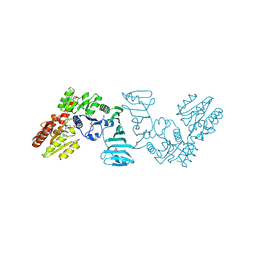 | | Crystal structure of SdgB (complexed with phosphate ions) | | Descriptor: | Glycosyl transferase, group 1 family protein, PHOSPHATE ION | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5NNS
 
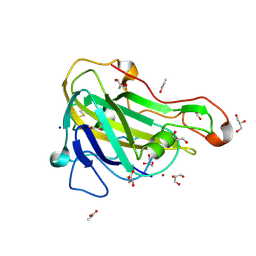 | | Crystal structure of HiLPMO9B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, COPPER (II) ION, ... | | Authors: | Dimarogona, M, Sandgren, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition.
FEBS J., 285, 2018
|
|
7C13
 
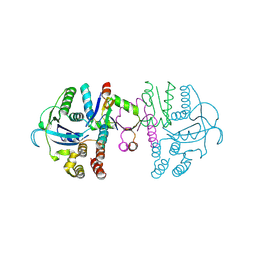 | | beta1 domain-swapped structure of monothiol cGrx1(C16S) | | Descriptor: | Glutaredoxin, Peptide methionine sulfoxide reductase MsrA | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
7C12
 
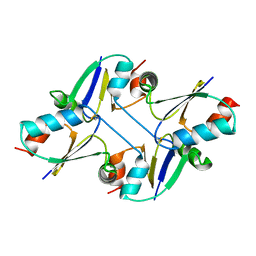 | | beta1 domain-swapped structure of monothiol cGrx1(C16S) | | Descriptor: | Glutaredoxin | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
7C10
 
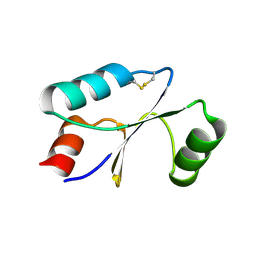 | | Dithiol cGrx1 | | Descriptor: | Glutaredoxin | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
2QKY
 
 | | complex structure of dipeptidyl peptidase IV and a oxadiazolyl ketone | | Descriptor: | 2-[(2-{(2S,4S)-2-[(R)-(5-tert-butyl-1,3,4-oxadiazol-2-yl)(hydroxy)methyl]-4-fluoropyrrolidin-1-yl}-2-oxoethyl)amino]-2-methylpropan-1-ol, Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) (Dipeptidyl peptidase 4 soluble form) (Dipeptidyl peptidase IV soluble form) | | Authors: | Kim, K.-H, Hong, S.Y, Koo, K.D, Lee, C.-S, Kim, G.T, Han, H.O. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis, SAR, and X-ray structure of novel potent DPPIV inhibitors: oxadiazolyl ketones.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6IJQ
 
 | | Solution structure of BCL-XL bound to P73-TAD peptide | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Tumor protein p73 | | Authors: | Yoon, M.-K, Ha, J.-H, Lee, M.-S, Chi, S.-W. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cytoplasmic pro-apoptotic function of the tumor suppressor p73 is mediated through a modified mode of recognition of the anti-apoptotic regulator Bcl-XL.
J. Biol. Chem., 293, 2018
|
|
5NBS
 
 | | Structural studies of a Glycoside Hydrolase Family 3 beta-glucosidase from the Model Fungus Neurospora crassa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Gudmundsson, M, Karkehabadi, S, Kaper, T, Sandgren, M. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies of a glycoside hydrolase family 3 beta-glucosidase from the model fungus Neurospora crassa.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
