7KUE
 
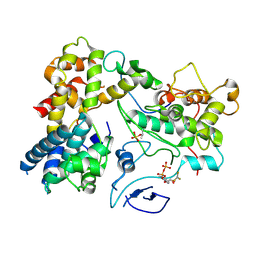 | | CryoEM structure of Yeast TFIIK (Kin28/Ccl1/Tfb3) Complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin CCL1, ... | | 著者 | van Eeuwen, T, Murakami, K, Li, T, Tsai, K.L. | | 登録日 | 2020-11-24 | | 公開日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of TFIIK for phosphorylation of CTD of RNA polymerase II.
Sci Adv, 7, 2021
|
|
1POO
 
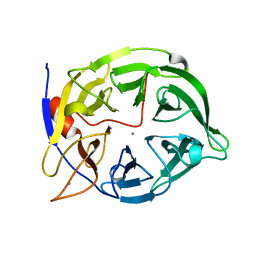 | | THERMOSTABLE PHYTASE FROM BACILLUS SP | | 分子名称: | CALCIUM ION, PROTEIN (PHYTASE) | | 著者 | Oh, B.H, Ha, N.C. | | 登録日 | 1999-04-16 | | 公開日 | 2000-04-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
7M2U
 
 | |
1QLG
 
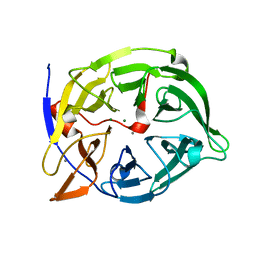 | | Crystal structure of phytase with magnesium from Bacillus amyloliquefaciens | | 分子名称: | 3-PHYTASE, CALCIUM ION, MAGNESIUM ION | | 著者 | Shin, S, Ha, N.-C, Oh, B.-H. | | 登録日 | 1999-08-31 | | 公開日 | 2000-02-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structures of a Novel, Thermostable Phytase in Partially and Fully Calcium-Loaded States
Nat.Struct.Biol., 7, 2000
|
|
7MEI
 
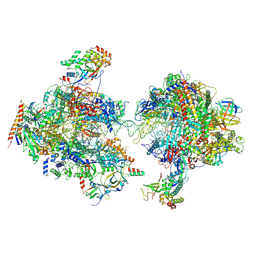 | | Composite structure of EC+EC | | 分子名称: | DNA (74-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Yang, C, Murakami, K. | | 登録日 | 2021-04-06 | | 公開日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MKA
 
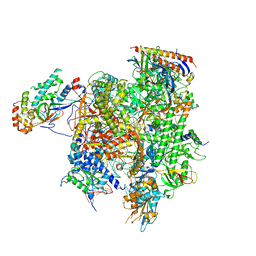 | | Structure of EC+EC (leading EC-focused) | | 分子名称: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Yang, C, Murakami, K. | | 登録日 | 2021-04-22 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-05-17 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MK9
 
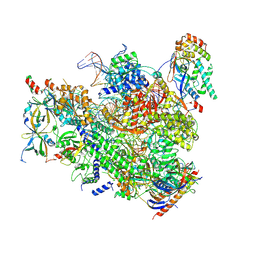 | |
6IY0
 
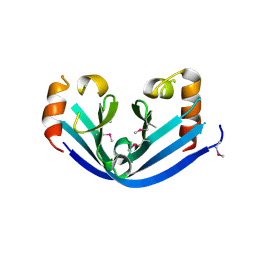 | |
6JQS
 
 | | Structure of Transcription factor, GerE | | 分子名称: | DNA-binding response regulator | | 著者 | Lee, J.H, Lee, C.W. | | 登録日 | 2019-04-01 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
5YCN
 
 | | Human PPARgamma ligand binding domain complexed with Lobeglitazone | | 分子名称: | (5S)-5-[[4-[2-[[6-(4-methoxyphenoxy)pyrimidin-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | 著者 | Jang, J.Y, Han, B.W. | | 登録日 | 2017-09-07 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPAR gamma.
Sci Rep, 8, 2018
|
|
5YCP
 
 | |
6A0H
 
 | |
6A0F
 
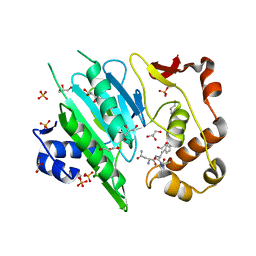 | |
5CEH
 
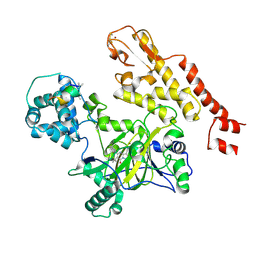 | | Structure of histone lysine demethylase KDM5A in complex with selective inhibitor | | 分子名称: | 7-oxo-5-phenyl-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, NICKEL (II) ION, ... | | 著者 | Kiefer, J.R, Vinogradova, M. | | 登録日 | 2015-07-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells.
Nat.Chem.Biol., 12, 2016
|
|
6A0I
 
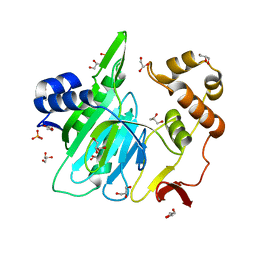 | |
6A0E
 
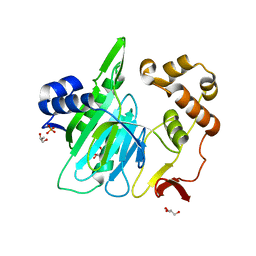 | |
4RHD
 
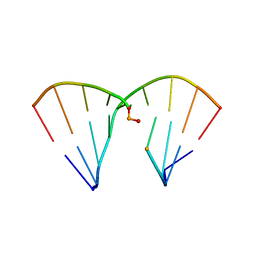 | | DNA Duplex with Novel ZP Base Pair | | 分子名称: | DNA 9mer novel P nucleobase, DNA 9mer novel Z nucleobase, MAGNESIUM ION | | 著者 | Zhang, W, Zhang, L, Benner, S, Huang, Z. | | 登録日 | 2014-10-01 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evolution of functional six-nucleotide DNA.
J.Am.Chem.Soc., 137, 2015
|
|
2A5E
 
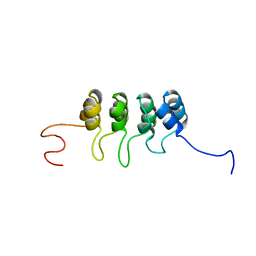 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | 分子名称: | TUMOR SUPPRESSOR P16INK4A | | 著者 | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | 登録日 | 1998-02-13 | | 公開日 | 1999-08-13 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
6KTN
 
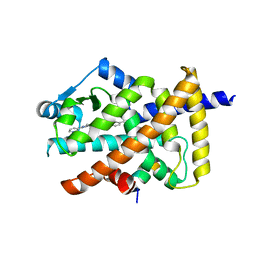 | | Human PPARgamma ligand-binding domain R288A mutant in complex with imatinib | | 分子名称: | 16-mer peptide from Nuclear receptor coactivator 1, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Peroxisome proliferator-activated receptor gamma | | 著者 | Jang, J.Y, Han, B.W. | | 登録日 | 2019-08-28 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.752 Å) | | 主引用文献 | Structural Basis for the Regulation of PPAR gamma Activity by Imatinib.
Molecules, 24, 2019
|
|
6KTM
 
 | |
2HZQ
 
 | |
2HZR
 
 | |
5XGW
 
 | |
5XGX
 
 | | Crystal structure of colwellia psychrerythraea strain 34H isoaspartyl dipeptidase E80Q mutant complexed with beta-isoaspartyl lysine | | 分子名称: | D-ASPARTIC ACID, D-LYSINE, Isoaspartyl dipeptidase, ... | | 著者 | Lee, J.H, Lee, C.W, Park, S.H. | | 登録日 | 2017-04-18 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal structure and functional characterization of an isoaspartyl dipeptidase (CpsIadA) from Colwellia psychrerythraea strain 34H.
PLoS ONE, 12, 2017
|
|
5Z8A
 
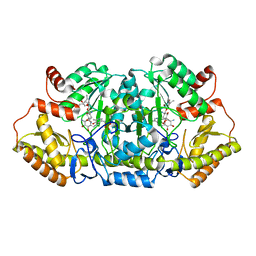 | | Crystal structure of GenB1 from Micromonospora echinospora in complex with JI-20A and PLP (external aldimine) | | 分子名称: | (2~{R},3~{R},4~{R},5~{R})-2-[(1~{S},2~{S},3~{R},4~{S},6~{R})-3-[(2~{R},3~{R},4~{R},5~{S},6~{R})-6-(aminomethyl)-3-azany l-4,5-bis(oxidanyl)oxan-2-yl]oxy-4,6-bis(azanyl)-2-oxidanyl-cyclohexyl]oxy-5-methyl-4-(methylamino)oxane-3,5-diol, C-6' aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Hong, S.K, Cha, S.S. | | 登録日 | 2018-01-31 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.398 Å) | | 主引用文献 | Complete reconstitution of the diverse pathways of gentamicin B biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
