7VCJ
 
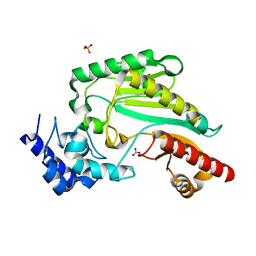 | | Arginine kinase H227A from Daphnia magna | | Descriptor: | Arginine kinase, NITRATE ION, PHOSPHATE ION | | Authors: | Kim, D.S, Jang, K, Kim, W.S, Kim, Y.J, Park, J.H. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability.
Molecules, 27, 2022
|
|
7UPH
 
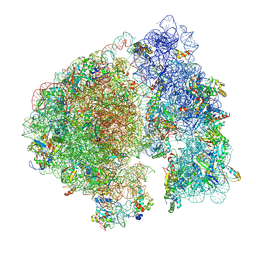 | | Structure of a ribosome with tethered subunits | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Kim, D.S, Watkins, A, Bidstrup, E, Lee, J, Topkar, V.V, Kofman, C, Schwarz, K.J, Liu, Y, Pintilie, G, Roney, E, Das, R, Jewett, M.C. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Three-dimensional structure-guided evolution of a ribosome with tethered subunits.
Nat.Chem.Biol., 18, 2022
|
|
5D43
 
 | |
6KY2
 
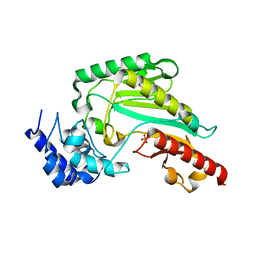 | | Crystal Structure of Arginine Kinase wild type from Daphnia magna | | Descriptor: | Arginine kinase, PHOSPHATE ION | | Authors: | Park, J.H, Rao, Z, Kim, S.Y, Kim, D.S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Insight into Structural Aspects of Histidine 284 of Daphnia magna Arginine Kinase.
Mol.Cells, 43, 2020
|
|
6KY3
 
 | | Structure of arginine kinase H284A mutant | | Descriptor: | ARGININE, Arginine kinase, PHOSPHATE ION, ... | | Authors: | Rao, Z, Park, J.H, Kim, S.Y, Kim, D.S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Insight into Structural Aspects of Histidine 284 of Daphnia magna Arginine Kinase.
Mol.Cells, 43, 2020
|
|
6L2U
 
 | | Soluble methane monooxygenase reductase FAD-binding domain from Methylosinus sporium. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methane monooxygenase | | Authors: | Park, J.H, Ha, S.C, Rao, Z, Yoo, H, Yoon, C, Kim, S.Y, Kim, D.S, Lee, S.J. | | Deposit date: | 2019-10-07 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of the electron transfer environment in the MMOR FAD-binding domain from Methylosinus sporium 5.
Dalton Trans, 50, 2021
|
|
7DGR
 
 | | Activity optimized supercomplex state2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGQ
 
 | | Activity optimized supercomplex state1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGZ
 
 | | Activity optimized complex I (closed form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DKF
 
 | | Activity optimized supercomplex state4 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DH0
 
 | | Activity optimized complex I (open form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
7DGS
 
 | | Activity optimized supercomplex state3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Jeon, T.J, Lee, S.G, Yoo, S.H, Ryu, J.H, Kim, D.S, Hyun, J.K, Kim, H.M, Ryu, S.E. | | Deposit date: | 2020-11-12 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | A Dynamic Substrate Pool Revealed by cryo-EM of a Lipid-Preserved Respiratory Supercomplex.
Antioxid.Redox Signal., 2022
|
|
6AHG
 
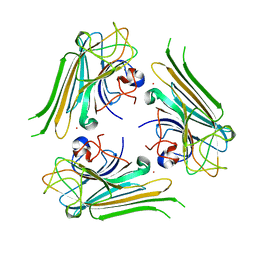 | | Trimeric structure of concanavalin A from Canavalia ensiformis | | Descriptor: | CADMIUM ION, CALCIUM ION, Concanavalin-A,Concanavalin-A | | Authors: | Park, J.H, Kim, D.S, Park, Y.R, Lee, S.J. | | Deposit date: | 2018-08-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Cadmium-substituted concanavalin A and its trimeric complexation
J. Microbiol. Biotechnol., 28(12), 2018
|
|
6XF4
 
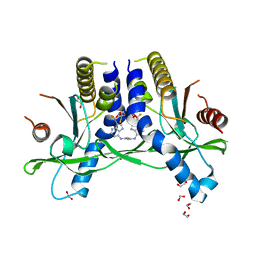 | | Crystal structure of STING REF variant in complex with E7766 | | Descriptor: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), 1,2-ETHANEDIOL, Stimulator of interferon genes protein | | Authors: | Chen, Y, Wang, J.Y, Kim, D.-S. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
6XF3
 
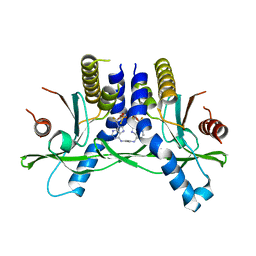 | | Crystal structure of STING in complex with E7766 | | Descriptor: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), Stimulator of interferon genes protein | | Authors: | Chen, Y, Wang, J.Y, Kim, D.-S. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
6GEZ
 
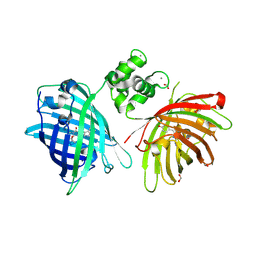 | | THE STRUCTURE OF TWITCH-2B N532F | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
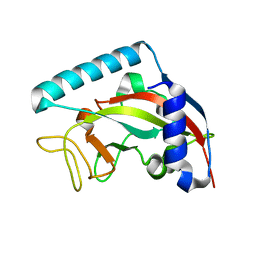
5NGO
 
 | |
6A6K
 
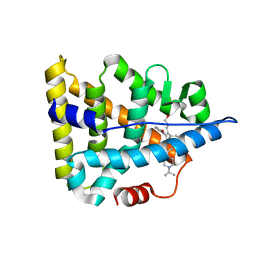 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | Descriptor: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
4JMK
 
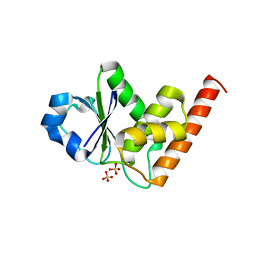 | | Structure of dusp8 | | Descriptor: | Dual specificity protein phosphatase 8, SULFATE ION | | Authors: | Jeong, D.G, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JMJ
 
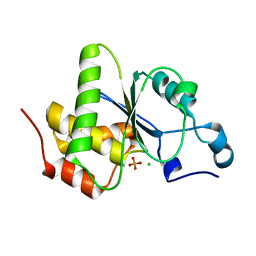 | | Structure of dusp11 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RNA/RNP complex-1-interacting phosphatase | | Authors: | Jeong, D.G, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KI9
 
 | | Crystal structure of the catalytic domain of human DUSP12 at 2.0 A resolution | | Descriptor: | Dual specificity protein phosphatase 12, PHOSPHATE ION | | Authors: | Jeon, T.J, Chien, P.N, Ku, B, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-05-02 | | Release date: | 2014-02-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JNB
 
 | | Crystal structure of the Catalytic Domain of Human DUSP12 | | Descriptor: | Dual specificity protein phosphatase 12, SULFATE ION | | Authors: | Jeon, T.J, Chien, P.N, Ku, B, Kim, S.J, Ryu, S.E. | | Deposit date: | 2013-03-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The family-wide structure and function of human dual-specificity protein phosphatases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6KNR
 
 | | Crystal structure of Estrogen-related receptor gamma ligand-binding domain with DN200699 | | Descriptor: | (E)-4-(1-(4-(1-cyclopropylpiperidin-4-yl)phenyl)-5-hydroxy-2-phenylpent-1-en-1-yl)phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Song, J, Cho, S.J. | | Deposit date: | 2019-08-07 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | An orally available inverse agonist of estrogen-related receptor gamma showed expanded efficacy for the radioiodine therapy of poorly differentiated thyroid cancer.
Eur.J.Med.Chem., 205, 2020
|
|
6GEL
 
 | | The structure of TWITCH-2B | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-26 | | Release date: | 2019-08-21 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
5UKG
 
 | |
