6YJ2
 
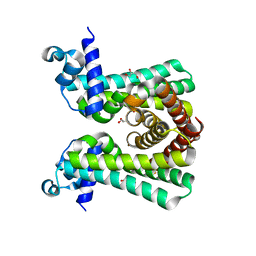 | | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism | | Descriptor: | GLYCEROL, Probable transcriptional regulatory protein (Probably TetR-family) | | Authors: | Keep, N.H, Pritchard, J.E, Sula, A, Cole, A.R, Kendall, S.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism
To be published
|
|
6YL2
 
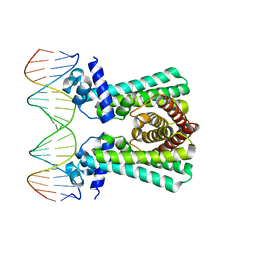 | | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*TP*AP*AP*TP*GP*AP*CP*GP*AP*TP*TP*AP*AP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*TP*TP*AP*AP*TP*CP*GP*TP*CP*AP*TP*TP*AP*AP*CP*GP*T)-3'), Probable transcriptional regulatory protein (Probably TetR-family) | | Authors: | Keep, N.H, Pritchard, J.E, Sula, A, Cole, A.R, Kendall, S.L. | | Deposit date: | 2020-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism
To be published
|
|
3MNL
 
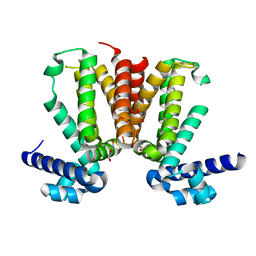 | | The crystal structure of KstR (Rv3574) from Mycobacterium tuberculosis H37Rv | | Descriptor: | TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY TETR-FAMILY), TRIETHYLENE GLYCOL | | Authors: | Gao, C, Bunker, R.D, ten Bokum, A, Kendall, S.L, Stoker, N.G, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-21 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J. Biol. Chem., 291, 2016
|
|
5CW8
 
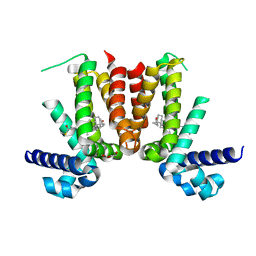 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with 3-oxo-4-cholesten-26-oyl-CoA | | Descriptor: | HTH-type transcriptional repressor KstR, S-{1-[5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,7-dihydroxy-6,6-dimethyl-3-oxido-8,12 -dioxo-2,4-dioxa-9,13-diaza-3lambda~5~-phosphapentadecan-15-yl} (2S,6R)-6-[(8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3-oxo-2,3,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta [a]phenanthren-17-yl]-2-methylheptanethioate (non-preferred name), TRIETHYLENE GLYCOL | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Casabon, I, Crowe, A.M, Baker, E.N, Eltis, L.D, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-27 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5CXG
 
 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with PEG | | Descriptor: | DI(HYDROXYETHYL)ETHER, HTH-type transcriptional repressor KstR, TRIETHYLENE GLYCOL | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1001 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5CXI
 
 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with 3-oxo-23,24-bisnorchol-4-en-22-oyl-CoA (4-BNC-CoA) | | Descriptor: | 3-oxo-23,24-bisnorchol-4-en-22-oyl-CoA, HTH-type transcriptional repressor KstR | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Casabon, I, Crowe, A.M, Baker, E.N, Eltis, L.D, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
7OMX
 
 | | Thermus sp. 2.9 DarT in complex with carba-NAD+ | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DarT domain-containing protein, THIOCYANATE ION | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMV
 
 | | Thermus sp. 2.9 DarT | | Descriptor: | CHLORIDE ION, DarT domain-containing protein, THIOCYANATE ION | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7ON0
 
 | | Thermus sp. 2.9 DarT in complex with ADP-ribosylated ssDNA | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*C)-3'), DarT domain-containing protein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMZ
 
 | |
7OMW
 
 | | Thermus sp. 2.9 DarT in complex with NAD+ | | Descriptor: | DarT domain-containing protein, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMY
 
 | | Thermus sp. 2.9 DarT in complex with carba-NAD+ and ssDNA | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DNA (5'-D(*AP*TP*GP*TP*C)-3'), DarT domain-containing protein, ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMU
 
 | | Thermosipho africanus DarTG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Macro domain-containing protein | | Authors: | Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
3ZXQ
 
 | |
3ZXO
 
 | | CRYSTAL STRUCTURE OF THE MUTANT ATP-BINDING DOMAIN OF MYCOBACTERIUM TUBERCULOSIS DOSS | | Descriptor: | ACETATE ION, GLYCEROL, REDOX SENSOR HISTIDINE KINASE RESPONSE REGULATOR DEVS, ... | | Authors: | Cho, H.Y, Cho, H.J, Kang, B.S. | | Deposit date: | 2011-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activation of ATP Binding for the Autophosphorylation of Doss, a Mycobacterium Tuberculosis Histidine Kinase Lacking an ATP-Lid Motif.
J.Biol.Chem., 288, 2013
|
|
1XKF
 
 | |
1Y5H
 
 | |
