6P63
 
 | |
8YTR
 
 | | The structure of Cu(II)-CopC from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, CopC domain-containing protein, DI(HYDROXYETHYL)ETHER | | Authors: | Kulikova, O.G, Solovieva, A.Y, Varfolomeeva, L.A, Dergousova, N.I, Nikolaeva, A.Y, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Cu(II)-CopC from Thioalkalivibrio paradoxus
To Be Published
|
|
8YTS
 
 | | The structure of the cytochrome c546/556 from Thioalkalivibrio paradoxus with unusual UV-Vis spectral features at atomic resolution | | Descriptor: | Cytochrome C, HEME C | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The structure of the cytochrome c546/556 from Thioalkalivibrio paradoxus with unusual UV-Vis spectral features at atomic resolution
To Be Published
|
|
8YU6
 
 | | The structure of thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q), activated by crystal soaking with 1mM CuCl2 and 1 mM sodium ascorbate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q), activated by crystal soaking with 1mM CuCl2 and 1 mM sodium ascorbate
To Be Published
|
|
8YQ4
 
 | | Structure of mBaoJin2 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Minyaev, M.E, Kuzmicheva, T.P, Vlaskina, A.V, Popov, V.O, Pyatkevich, K.D, Subach, F.V. | | Deposit date: | 2024-03-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mBaoJin2
To Be Published
|
|
8YRV
 
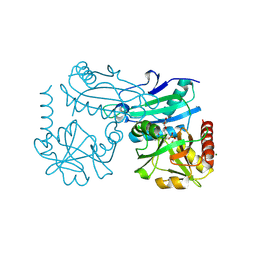 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, MAGNESIUM ION | | Authors: | Matyuta, I.O, Bakunova, A.K, Nikolaeva, A.Y, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis complexed with 3-aminooxypropionic acid
To Be Published
|
|
8YRT
 
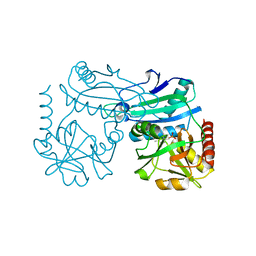 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis in the holo form obtained at pH 7.0 | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis in the holo form obtained at pH 7.0
To Be Published
|
|
8YTQ
 
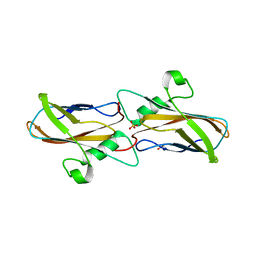 | | The structure of apoCopC from Thioalkalivibrio paradoxus | | Descriptor: | ACETATE ION, COPPER (II) ION, CopC domain-containing protein, ... | | Authors: | Kulikova, O.G, Solovieva, A.Y, Varfolomeeva, L.A, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of apoCopC from Thioalkalivibrio paradoxus
To Be Published
|
|
8YRU
 
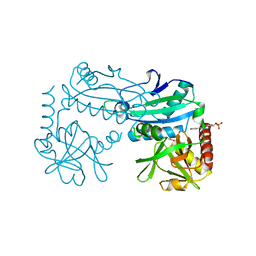 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (apo form) after 15 sec of soaking with phenylhydrazine | | Descriptor: | ACETATE ION, Aminotransferase class IV, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (apo form) after 15 sec of soaking with phenylhydrazine
To Be Published
|
|
8YU5
 
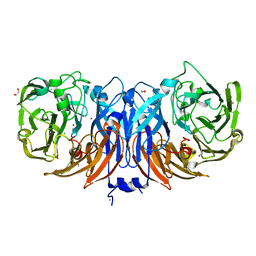 | | The structure of non-activated thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of non-activated thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q)
To Be Published
|
|
2ACS
 
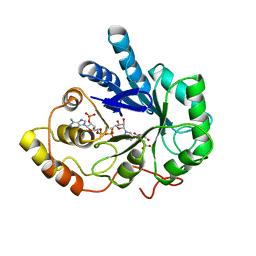 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
2ACR
 
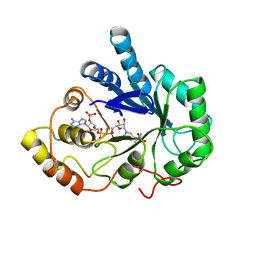 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | ALDOSE REDUCTASE, CACODYLATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
3RKH
 
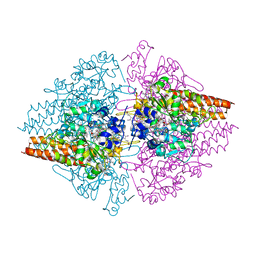 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite (full occupancy) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-04-18 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2ACQ
 
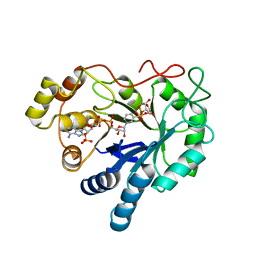 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
8G4Z
 
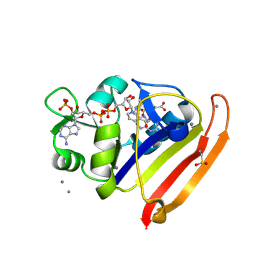 | | E. coli DHFR complex with NADP+ and folate: EF-X off model by Laue diffraction (no electric field) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Brookner, D.E, Klureza, M.A, Hekstra, D.R. | | Deposit date: | 2023-02-10 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | E. coli DHFR complex with NADP+ and folate: EF-X off model by Laue diffraction (no electric field)
To Be Published
|
|
8G50
 
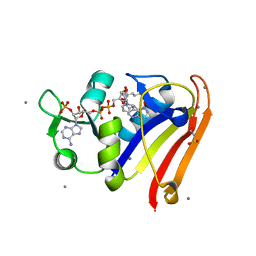 | | E. coli DHFR complex with NADP+ and folate: EF-X excited state model by Laue diffraction (electric field along b axis; 8-fold extrapolation of structure factor differences) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Brookner, D.E, Klureza, M.A, Hekstra, D.R. | | Deposit date: | 2023-02-10 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | E. coli DHFR complex with NADP+ and folate: EF-X excited state model by Laue diffraction (electric field along b axis; 8-fold extrapolation of structure factor differences)
To Be Published
|
|
8FRF
 
 | |
8FRE
 
 | |
7RIN
 
 | |
7RG4
 
 | | Importin alpha2 in complex with p50 NLS | | Descriptor: | Importin subunit alpha-1, Isoform 3 of Nuclear factor NF-kappa-B p105 subunit | | Authors: | Smith, K.M, Tsimbalyuk, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
7RG5
 
 | | Importin alpha3 in complex with p50 NLS | | Descriptor: | Importin subunit alpha-3, Isoform 3 of Nuclear factor NF-kappa-B p105 subunit | | Authors: | Smith, K.M, Tsimbalyuk, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
8GJW
 
 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJY
 
 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJX
 
 | | Structure of the human STING receptor bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Morehouse, B.R, Li, Y, Slavik, K.M, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
7R86
 
 | | Structure of mouse BAI1 (ADGRB1) in complex with mouse Nogo receptor (RTN4R) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINOBENZOIC ACID, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
