7APQ
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-(1,3-benzothiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Kolos, M.J, Pomplun, S, Riess, B, Purder, P, Voll, M.A, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
7APT
 
 | | The Fk1 domain of FKBP51 in complex with ((1S,5S,6R)-10-((3,5-dichlorophenyl)sulfonyl)-2-oxo-5-vinyl-3,10-diazabicyclo[4.3.1]decan-3-yl)acetic acid | | Descriptor: | 2-[(1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-2-oxidanylidene-3,10-diazabicyclo[4.3.1]decan-3-yl]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Kolos, M.J, Pomplun, S, Riess, B, Purder, P, Voll, M.A, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
7APW
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, A.M, Kolos, J.M, Pomplun, S, Riess, B, Purder, P, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-20 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
8GJV
 
 | |
8GW6
 
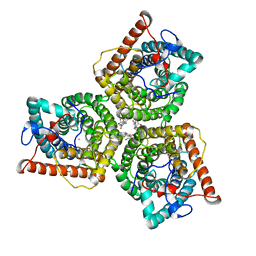 | | AtSLAC1 6D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2022-09-16 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8GW7
 
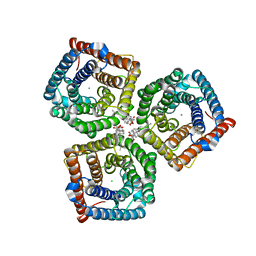 | | AtSLAC1 6D mutant in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2022-09-16 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
7S24
 
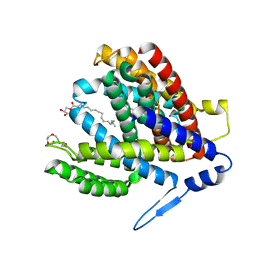 | | Crystal structure of the Na+/H+ antiporter NhaA at pH 6.5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Na(+)/H(+) antiporter NhaA, PENTAETHYLENE GLYCOL | | Authors: | Drew, D, Brock, J, Uzdavinys, P, Matsuoka, R. | | Deposit date: | 2021-09-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Na + /H + antiporter NhaA at active pH reveals the mechanistic basis for pH sensing.
Nat Commun, 13, 2022
|
|
8J1E
 
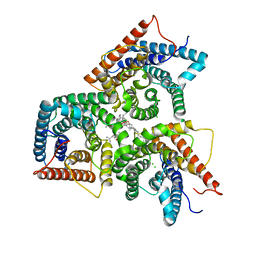 | | AtSLAC1 in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J0J
 
 | | AtSLAC1 8D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
4E90
 
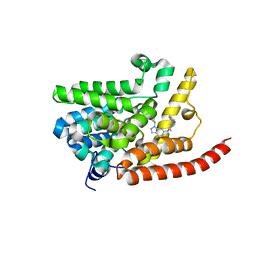 | | Human phosphodiesterase 9 in complex with inhibitors | | Descriptor: | 6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyran-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-03-20 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
4G2L
 
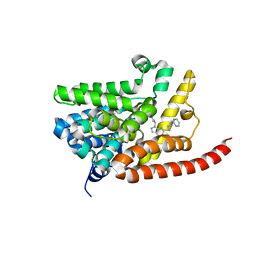 | | Human PDE9 in complex with selective compound | | Descriptor: | 1-cyclopentyl-6-{(1R)-1-[3-(pyrimidin-2-yl)azetidin-1-yl]ethyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-07-12 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
4G2J
 
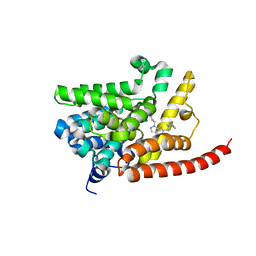 | | Human pde9 in complex with selective compound | | Descriptor: | 1-cyclopentyl-6-[(1R)-1-(3-phenoxyazetidin-1-yl)ethyl]-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-07-12 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
1PAX
 
 | |
4IMK
 
 | | Uncrossed Fab binding to human Angiopoietin 2 | | Descriptor: | GLYCEROL, Heavy Chain, Light Chain, ... | | Authors: | Fenn, S, Schiller, C, Griese, J.J, Hopfner, K.-P, Kettenberger, H. | | Deposit date: | 2013-01-03 | | Release date: | 2013-04-17 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal Structure of an Anti-Ang2 CrossFab Demonstrates Complete Structural and Functional Integrity of the Variable Domain.
Plos One, 8, 2013
|
|
4IML
 
 | | CrossFab binding to human Angiopoietin 2 | | Descriptor: | Crossed heavy chain (VH-Ckappa), Crossed light chain (VL-CH1), GLYCEROL | | Authors: | Fenn, S, Schiller, C, Griese, J, Hopfner, K.-P, Kettenberger, H. | | Deposit date: | 2013-01-03 | | Release date: | 2013-04-17 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Crystal Structure of an Anti-Ang2 CrossFab Demonstrates Complete Structural and Functional Integrity of the Variable Domain.
Plos One, 8, 2013
|
|
7ZO9
 
 | | cryo-EM structure of CGT ABC transporter in vanadate trapped state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, VANADATE ION | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZNU
 
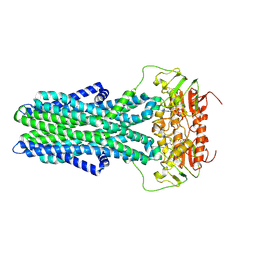 | | cryo-EM structure of CGT ABC transporter in detergent micelle | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, VANADATE ION | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZO8
 
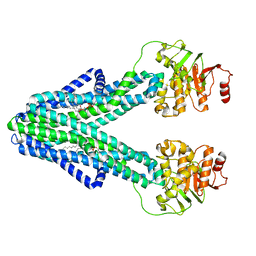 | | cryo-EM structure of CGT ABC transporter in nanodisc apo state | | Descriptor: | Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, DIUNDECYL PHOSPHATIDYL CHOLINE | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZOA
 
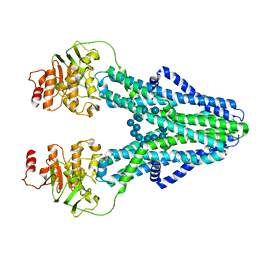 | | cryo-EM structure of CGT ABC transporter in presence of CBG substrate | | Descriptor: | Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, Cyclooctadecakis-(1-2)-(beta-D-glucopyranose) | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3PAX
 
 | |
2PAW
 
 | | THE CATALYTIC FRAGMENT OF POLY(ADP-RIBOSE) POLYMERASE | | Descriptor: | POLY(ADP-RIBOSE) POLYMERASE | | Authors: | Ruf, A, Schulz, G.E. | | Deposit date: | 1997-11-25 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling.
Biochemistry, 37, 1998
|
|
2PAX
 
 | |
7D9N
 
 | |
4PAX
 
 | |
2JSA
 
 | |
