2BT6
 
 | | Ru(bpy)2(mbpy)-Modified Bovine Adrenodoxin | | Descriptor: | (4'-METHYL-2,2'BIPYRIDINE)BIS(2,2'-BIPYRIDINE), ADRENODOXIN 1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Halavaty, A, Mueller, J.J, Contzen, J, Jung, C, Hannemann, F, Bernhardt, R, Galander, M, Lendzian, F, Heinemann, U. | | Deposit date: | 2005-05-26 | | Release date: | 2006-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Light-Induced Reduction of Bovine Adrenodoxin Via the Covalently Bound Ruthenium(II) Bipyridyl Complex: Intramolecular Electron Transfer and Crystal Structure.
Biochemistry, 45, 2006
|
|
384D
 
 | |
329D
 
 | |
382D
 
 | |
383D
 
 | |
1AKD
 
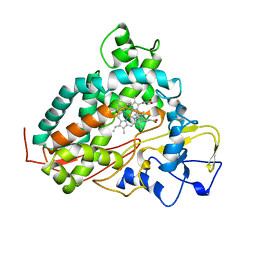 | | CYTOCHROME P450CAM FROM PSEUDOMONAS PUTIDA, COMPLEXED WITH 1S-CAMPHOR | | Descriptor: | CAMPHOR, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Schlichting, I, Jung, C, Schulze, H. | | Deposit date: | 1997-05-16 | | Release date: | 1997-11-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cytochrome P-450cam complexed with the (1S)-camphor enantiomer.
FEBS Lett., 415, 1997
|
|
6MDR
 
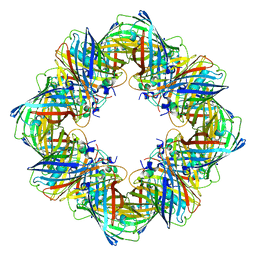 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
1J90
 
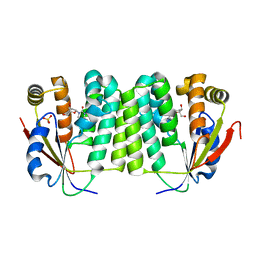 | | Crystal Structure of Drosophila Deoxyribonucleoside Kinase | | Descriptor: | 2'-DEOXYCYTIDINE, Deoxyribonucleoside kinase, SULFATE ION | | Authors: | Johansson, K, Ramaswamy, S, Ljungkrantz, C, Knecht, W, Piskur, J, Munch-Petersen, B, Eriksson, S, Eklund, H. | | Deposit date: | 2001-05-23 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for substrate specificities of cellular deoxyribonucleoside kinases.
Nat.Struct.Biol., 8, 2001
|
|
2OCP
 
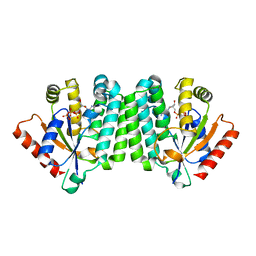 | | Crystal Structure of Human Deoxyguanosine Kinase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxyguanosine kinase | | Authors: | Johansson, K, Ramaswamy, S, Ljungkrantz, C, Knecht, W, Piskur, J, Munch-Petersen, B, Eriksson, S, Eklund, H. | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Substrate Specificities of Cellular Deoxyribonucleoside Kinases.
Nat.Struct.Biol., 8, 2001
|
|
7JTZ
 
 | | Yeast Glo3 GAP domain | | Descriptor: | ADP-ribosylation factor GTPase-activating protein GLO3, GLYCEROL, ZINC ION | | Authors: | Xie, B, Jackson, L.P. | | Deposit date: | 2020-08-18 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Glo3 GAP crystal structure supports the molecular niche model for ArfGAPs in COPI coats.
Adv Biol Regul, 79, 2021
|
|
3AQ3
 
 | |
3AQ4
 
 | |
3AQ2
 
 | |
8S86
 
 | | human PLD3 homodimer structure | | Descriptor: | 5'-3' exonuclease PLD3 | | Authors: | Lammens, K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Lysosomal endonuclease RNase T2 and PLD exonucleases cooperatively generate RNA ligands for TLR7 activation.
Immunity, 57, 2024
|
|
9GMO
 
 | |
7ZR1
 
 | | Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS (composite structure) | | Descriptor: | DH domain-containing protein, Double-strand break repair protein, FHA domain-containing protein, ... | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-05-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
7ZQY
 
 | | Chaetomium thermophilum Rad50 Zn hook | | Descriptor: | DH domain-containing protein, ZINC ION | | Authors: | Lammens, K, Rotheneder, M, Stakyte, K. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
8BAH
 
 | | Human Mre11-Nbs1 complex | | Descriptor: | Double-strand break repair protein MRE11, MANGANESE (II) ION, Nibrin | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-10-11 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
6LVP
 
 | |
6LVO
 
 | |
1AVG
 
 | |
8I4O
 
 | | Design of a split green fluorescent protein for sensing and tracking an beta-amyloid | | Descriptor: | Beta-amyloid, Split Green flourescent protein | | Authors: | Taegeun, Y, Jinsu, L, Jungmin, Y, Jungmin, C, Wondo, H, Song, J.J, Haksung, K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering of a Fluorescent Protein for a Sensing of an Intrinsically Disordered Protein through Transition in the Chromophore State.
Jacs Au, 3, 2023
|
|
