2MBV
 
 | | LMO4-LIM2 in complex with DEAF1 (404-418) | | Descriptor: | Fusion protein of LIM domain transcription factor LMO4 (77-147) and DEAF1 (404-418), ZINC ION | | Authors: | Joseph, S, Matthews, J.M, Kwan, A.H.-Y, Mackay, J.P, Cubeddu, L, Foo, P. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterisation of LIM-only protein 4 (LMO4) in complex with Deformed Epidermal Autoregulatory Factor-1 (DEAF1)
To be Published
|
|
8ZVY
 
 | | Alpha-Synuclein with H2a-H2b dimer complex structure. | | Descriptor: | CHLORIDE ION, Histone H2B 1.1,Histone H2A type 1, Isoform 1 of Alpha-synuclein | | Authors: | Padavattan, S, Jos, S, Kambaru, A. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and functional insights into the nuclear role of Parkinson's disease-associated alpha-synuclein as a histone chaperone.
Commun Biol, 8, 2025
|
|
8KDX
 
 | |
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | Authors: | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4LR6
 
 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | Descriptor: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
6U0D
 
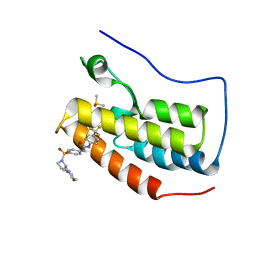 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0590 | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, N-[4-({(2S)-2-[(morpholin-4-yl)methyl]pyrrolidin-1-yl}sulfonyl)phenyl]-N'-[4-(trifluoromethyl)phenyl]urea | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Discovery, X-ray Crystallography, and Anti-inflammatory Activity of Bromodomain-containing Protein 4 (BRD4) BD1 Inhibitors Targeting a Distinct New Binding Site
J. Med. Chem., 2022
|
|
1E3K
 
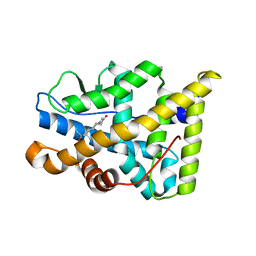 | | Human Progesteron Receptor Ligand Binding Domain in complex with the ligand metribolone (R1881) | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, PROGESTERONE RECEPTOR | | Authors: | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Egner, U, Carrondo, M.A. | | Deposit date: | 2000-06-19 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
1E3G
 
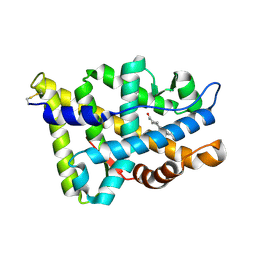 | | Human Androgen Receptor Ligand Binding in complex with the ligand metribolone (R1881) | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, ANDROGEN RECEPTOR | | Authors: | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Ruff, M, Egner, U, Carrondo, M.A. | | Deposit date: | 2000-06-14 | | Release date: | 2001-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
6UWU
 
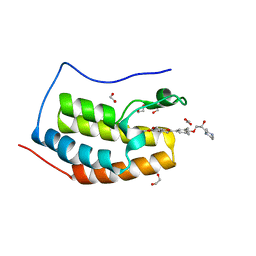 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0516 | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[(2R)-2-hydroxy-3-(4-methylpiperazin-1-yl)propoxy]-3,5-dimethylphenyl}-5,7-dimethoxy-4H-1-benzopyran-4-one, Bromodomain-containing protein 4 | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-11-05 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Orally Bioavailable Chromone Derivatives as Potent and Selective BRD4 Inhibitors: Scaffold Hopping, Optimization, and Pharmacological Evaluation.
J.Med.Chem., 63, 2020
|
|
2GV3
 
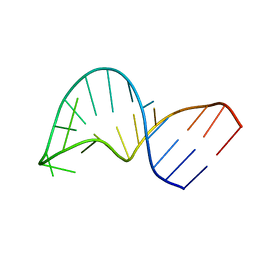 | | Translocation of a tRNA with an extended anticodon through the ribosome | | Descriptor: | 5'-R(*GP*GP*CP*CP*AP*GP*AP*CP*UP*CP*CP*CP*GP*AP*AP*UP*CP*UP*GP*GP*CP*C)-3' | | Authors: | Phelps, S.S, Gaudin, C, Yoshizawa, S, Benitez, C, Fourmy, D, Joseph, S. | | Deposit date: | 2006-05-02 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Translocation of a tRNA with an Extended Anticodon Through the Ribosome.
J.Mol.Biol., 360, 2006
|
|
2UXC
 
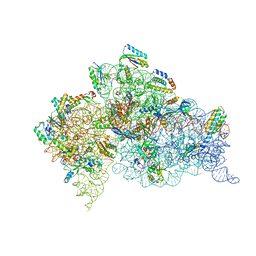 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA UCGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACGA, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
2UXB
 
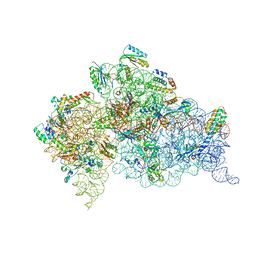 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA GGGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT GGGU, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACCC, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit.
RNA, 13, 2007
|
|
2UXD
 
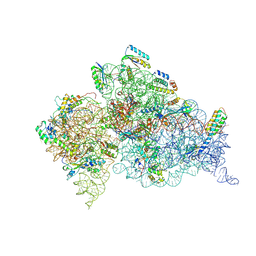 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON CCCG, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
7WHC
 
 | | Crystal structure of SARS-CoV-2 3CLpro catalytic domain | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Shin, D.H, Jo, S.R. | | Deposit date: | 2021-12-30 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Dimerization Tendency of 3CLpros of Human Coronaviruses Based on the X-ray Crystal Structure of the Catalytic Domain of SARS-CoV-2 3CLpro.
Int J Mol Sci, 23, 2022
|
|
6BV2
 
 | | Crystal structure of porcine aminopeptidase-N with Isoleucine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV4
 
 | | Crystal structure of porcine aminopeptidase-N with Methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BUY
 
 | | Crystal structure of porcine aminopeptidase-N with Glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-11 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV0
 
 | | Crystal structure of porcine aminopeptidase-N with Arginine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV1
 
 | | Crystal structure of porcine aminopeptidase-N with Aspartic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV3
 
 | | Crystal structure of porcine aminopeptidase-N with Leucine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
7UI6
 
 | |
1UIY
 
 | | Crystal Structure of Enoyl-CoA Hydratase from Thermus Thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Enoyl-CoA Hydratase, GLYCEROL | | Authors: | Bagautdinov, B, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-24 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
1I2U
 
 | | NMR SOLUTION STRUCTURES OF ANTIFUNGAL HELIOMICIN | | Descriptor: | DEFENSIN HELIOMICIN | | Authors: | Lamberty, M, Caille, A, Landon, C, Tassin-Moindrot, S, Hetru, C, Bulet, P, Vovelle, F. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities.
Biochemistry, 40, 2001
|
|
8G1A
 
 | | Cryo-EM structure of Nav1.7 with CBD | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-07-05 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cannabidiol inhibits Na v channels through two distinct binding sites.
Nat Commun, 14, 2023
|
|
6JQ8
 
 | |
