1HXH
 
 | | COMAMONAS TESTOSTERONI 3BETA/17BETA HYDROXYSTEROID DEHYDROGENASE | | Descriptor: | 3BETA/17BETA-HYDROXYSTEROID DEHYDROGENASE | | Authors: | Benach, J, Filling, C, Oppermann, U.C.T, Roversi, P, Bricogne, G, Berndt, K.D, Jornvall, H, Ladenstein, R. | | Deposit date: | 2001-01-15 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structure of Bacterial 3beta/17beta-Hydroxysteroid Dehydrogenase at 1.2 A Resolution: A Model for
Multiple Steroid Recognition
Biochemistry, 41, 2002
|
|
2YAD
 
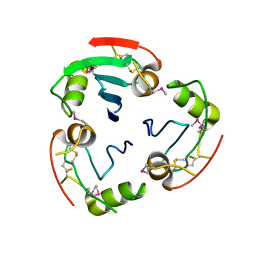 | | BRICHOS domain of Surfactant protein C precursor protein | | Descriptor: | SURFACTANT PROTEIN C BRICHOS DOMAIN | | Authors: | Askarieh, G, Siponen, M.I, Willander, H, Landreh, M, Westermark, P, Nordling, K, Keranen, H, Hermansson, E, Hamvas, A, Nogee, L.M, Bergman, T, Saenz, A, Casals, C, Aqvist, J, Jornvall, H, Presto, J, Johansson, J, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Nordlund, P, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, van den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Berglund, H, Knight, S.D. | | Deposit date: | 2011-02-18 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Resolution Structure of a Bricos Domain and its Implications for Anti-Amyloid Chaperone Activity on Lung Surgactant Protein C.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1G66
 
 | | ACETYLXYLAN ESTERASE AT 0.90 ANGSTROM RESOLUTION | | Descriptor: | ACETYL XYLAN ESTERASE II, GLYCEROL, SULFATE ION | | Authors: | Ghosh, D, Sawicki, M, Lala, P, Erman, M, Pangborn, W, Eyzaguirre, J, Gutierrez, R, Jornvall, H, Thiel, D.J. | | Deposit date: | 2000-11-03 | | Release date: | 2001-01-17 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Multiple conformations of catalytic serine and histidine in acetylxylan esterase at 0.90 A.
J.Biol.Chem., 276, 2001
|
|
1GQ8
 
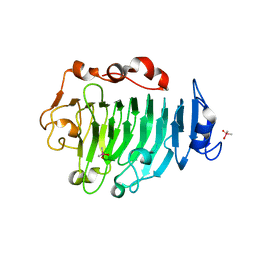 | | Pectin methylesterase from Carrot | | Descriptor: | CACODYLATE ION, PECTINESTERASE | | Authors: | Johansson, K, El-Ahmad, M, Friemann, R, Jornvall, H, Markovic, O, Eklund, H. | | Deposit date: | 2001-11-20 | | Release date: | 2002-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Plant Pectin Methylesterase
FEBS Lett., 514, 2002
|
|
2AXE
 
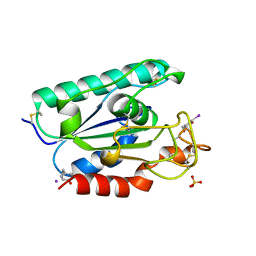 | | IODINATED COMPLEX OF ACETYL XYLAN ESTERASE AT 1.80 ANGSTROMS | | Descriptor: | ACETYL XYLAN ESTERASE, SULFATE ION | | Authors: | Ghosh, D, Erman, M, Sawicki, M.W, Lala, P, Weeks, D.R, Li, N, Pangborn, W, Thiel, D.J, Jornvall, H, Eyzaguirre, J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Determination of a protein structure by iodination: the structure of iodinated acetylxylan esterase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2ESY
 
 | | Structure and influence on stability and activity of the N-terminal propetide part of lung surfactant protein C | | Descriptor: | lung surfactant protein C | | Authors: | Li, J, Liepinsh, E, Almlen, A, Thyberg, J, Curstedt, T, Jornvall, H, Johansson, J. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure and influence on stability and activity of the N-terminal propeptide part of lung surfactant protein C
Febs J., 273, 2006
|
|
2MFZ
 
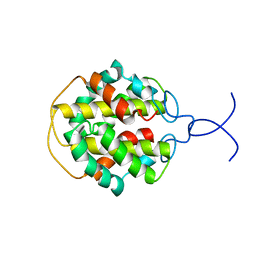 | | NMR structure of C-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Andersson, M, Chen, G, Landreh, M, Nordling, K, Kronqvist, N, Westermark, P, Jornvall, H, Knight, S, Ridderstrale, Y, Holm, L, Meng, Q, Chesler, M, Johansson, J, Rising, A. | | Deposit date: | 2013-10-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Carbonic Anhydrase Generates CO2 and H+ That Drive Spider Silk Formation Via Opposite Effects on the Terminal Domains
Plos Biol., 12, 2014
|
|
1A4S
 
 | | BETAINE ALDEHYDE DEHYDROGENASE FROM COD LIVER | | Descriptor: | BETAINE ALDEHYDE DEHYDROGENASE | | Authors: | Johansson, K, El Ahmad, M, Hjelmqvist, L, Ramaswamy, S, Jornvall, H, Eklund, H. | | Deposit date: | 1998-02-03 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of betaine aldehyde dehydrogenase at 2.1 A resolution.
Protein Sci., 7, 1998
|
|
1BPW
 
 | | BETAINE ALDEHYDE DEHYDROGENASE FROM COD LIVER | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (ALDEHYDE DEHYDROGENASE) | | Authors: | Johansson, K, El Ahmad, M, Ramaswamy, S, Hjelmqvist, L, Jornvall, H, Eklund, H. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of betaine aldehyde dehydrogenase at 2.1 A resolution.
Protein Sci., 7, 1998
|
|
1BK4
 
 | | CRYSTAL STRUCTURE OF RABBIT LIVER FRUCTOSE-1,6-BISPHOSPHATASE AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEIN (FRUCTOSE-1,6-BISPHOSPHATASE), SULFATE ION | | Authors: | Ghosh, D, Weeks, C.M, Erman, M, Roszak, A.W, Kaiser, R, Jornvall, H. | | Deposit date: | 1998-07-14 | | Release date: | 1998-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of rabbit liver fructose 1,6-bisphosphatase at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BS9
 
 | | ACETYLXYLAN ESTERASE FROM P. PURPUROGENUM REFINED AT 1.10 ANGSTROMS | | Descriptor: | ACETYL XYLAN ESTERASE, SULFATE ION | | Authors: | Ghosh, D, Erman, M, Sawicki, M.W, Lala, P, Weeks, D.R, Li, N, Pangborn, W, Thiel, D.J, Jornvall, H, Eyzaguirre, J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Determination of a protein structure by iodination: the structure of iodinated acetylxylan esterase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7ADH
 
 | |
6ADH
 
 | |
5ADH
 
 | |
8ADH
 
 | |
5NDA
 
 | | NMR Structural Characterisation of Pharmaceutically Relevant Proteins Obtained Through a Novel Recombinant Production: The Case of The Pulmonary Surfactant Polypeptide C Analogue rSP-C33Leu. | | Descriptor: | rSP-C33Leu -RECOMBINANT PULMONARY SURFACTANT-ASSOCIATED POLYPEPTIDE C ANALOGUE- | | Authors: | Venturi, L, Pioselli, B, Johansson, J, Rising, A, Kronqvist, N, Nordling, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Efficient protein production inspired by how spiders make silk.
Nat Commun, 8, 2017
|
|
2OHX
 
 | |
1ADC
 
 | | CRYSTALLOGRAPHIC STUDIES OF ISOSTERIC NAD ANALOGUES BOUND TO ALCOHOL DEHYDROGENASE: SPECIFICITY AND SUBSTRATE BINDING IN TWO TERNARY COMPLEXES | | Descriptor: | 5-BETA-D-RIBOFURANOSYLPICOLINAMIDE ADENINE-DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, ETHANOL, ... | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-12-13 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
Biochemistry, 33, 1994
|
|
1ADB
 
 | | CRYSTALLOGRAPHIC STUDIES OF ISOSTERIC NAD ANALOGUES BOUND TO ALCOHOL DEHYDROGENASE: SPECIFICITY AND SUBSTRATE BINDING IN TWO TERNARY COMPLEXES | | Descriptor: | 5-BETA-D-RIBOFURANOSYLNICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, ETHANOL, ... | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-12-13 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
Biochemistry, 33, 1994
|
|
4FBS
 
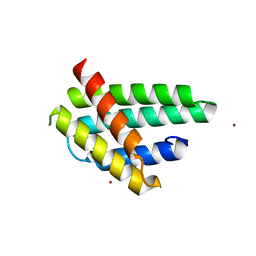 | | Structure of monomeric NT from Euprosthenops australis Major Ampullate Spidroin 1 (MaSp1) | | Descriptor: | BROMIDE ION, Major ampullate spidroin 1 | | Authors: | Askarieh, G, Hedhammar, M, Rising, A, Johansson, J, Knight, S.D. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | pH-Dependent Dimerization of Spider Silk N-Terminal Domain Requires Relocation of a Wedged Tryptophan Side Chain
J.Mol.Biol., 2012
|
|
1AXG
 
 | | CRYSTAL STRUCTURE OF THE VAL203->ALA MUTANT OF LIVER ALCOHOL DEHYDROGENASE COMPLEXED WITH COFACTOR NAD AND INHIBITOR TRIFLUOROETHANOL SOLVED TO 2.5 ANGSTROM RESOLUTION | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Colby, T.D, Chin, J.K, Bahnson, B.J, Goldstein, B.M, Klinman, J.P. | | Deposit date: | 1997-10-15 | | Release date: | 1998-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A link between protein structure and enzyme catalyzed hydrogen tunneling.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1ADG
 
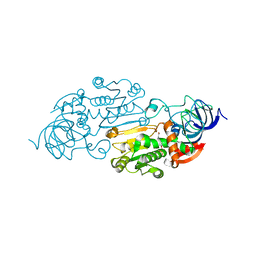 | | CRYSTALLOGRAPHIC STUDIES OF TWO ALCOHOL DEHYDROGENASE-BOUND ANALOGS OF THIAZOLE-4-CARBOXAMIDE ADENINE DINUCLEOTIDE (TAD), THE ACTIVE ANABOLITE OF THE ANTITUMOR AGENT TIAZOFURIN | | Descriptor: | ALCOHOL DEHYDROGENASE, BETA-METHYLENE-SELENAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, ZINC ION | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Marquez, V.E, Carrell, H.L, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-10-18 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of two alcohol dehydrogenase-bound analogues of thiazole-4-carboxamide adenine dinucleotide (TAD), the active anabolite of the antitumor agent tiazofurin.
Biochemistry, 33, 1994
|
|
1ADF
 
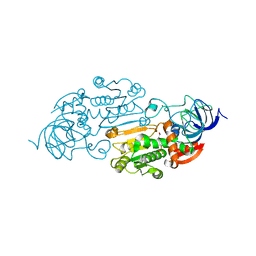 | | CRYSTALLOGRAPHIC STUDIES OF TWO ALCOHOL DEHYDROGENASE-BOUND ANALOGS OF THIAZOLE-4-CARBOXAMIDE ADENINE DINUCLEOTIDE (TAD), THE ACTIVE ANABOLITE OF THE ANTITUMOR AGENT TIAZOFURIN | | Descriptor: | ALCOHOL DEHYDROGENASE, BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, ZINC ION | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Marquez, V.E, Carrell, H.L, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-10-18 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of two alcohol dehydrogenase-bound analogues of thiazole-4-carboxamide adenine dinucleotide (TAD), the active anabolite of the antitumor agent tiazofurin.
Biochemistry, 33, 1994
|
|
1CLE
 
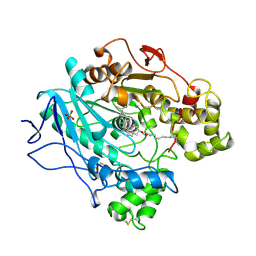 | | STRUCTURE OF UNCOMPLEXED AND LINOLEATE-BOUND CANDIDA CYLINDRACEA CHOLESTEROL ESTERASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL ESTERASE, ... | | Authors: | Ghosh, D. | | Deposit date: | 1995-02-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of uncomplexed and linoleate-bound Candida cylindracea cholesterol esterase.
Structure, 3, 1995
|
|
3GRX
 
 | | NMR STRUCTURE OF ESCHERICHIA COLI GLUTAREDOXIN 3-GLUTATHIONE MIXED DISULFIDE COMPLEX, 20 STRUCTURES | | Descriptor: | GLUTAREDOXIN 3, GLUTATHIONE | | Authors: | Nordstrand, K, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-30 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism.
J.Mol.Biol., 286, 1999
|
|
