8SQH
 
 | | SpRY-Cas9:gRNA complex targeting TTC PAM DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-05-04 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8SRS
 
 | | SpRY-Cas9:gRNA complex targeting TAC PAM DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-05-06 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8SPQ
 
 | | SpRY-Cas9:gRNA complex targeting TGG PAM DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8G2O
 
 | | Fab structure - Anti-ApoE-7C11 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, anti-ApoE-7C11 heavy chain, anti-ApoE-7C11 light chain | | Authors: | Marino, C, Arboleda-Velasquez, J.F. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | APOE Christchurch-mimetic therapeutic antibody reduces APOE-mediated toxicity and tau phosphorylation.
Alzheimers Dement, 20, 2024
|
|
1GJY
 
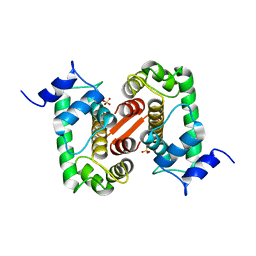 | | The X-ray structure of the Sorcin Calcium Binding Domain (SCBD) provides insight into the phosphorylation and calcium dependent processess | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Ilari, A, Johnson, K.A, Nastopoulos, V, Tsernoglou, D, Chiancone, E. | | Deposit date: | 2001-08-06 | | Release date: | 2002-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Sorcin Calcium Binding Domain Provides a Model of Ca(2+)-Dependent Processes in the Full-Length Protein
J.Mol.Biol., 317, 2002
|
|
1Q53
 
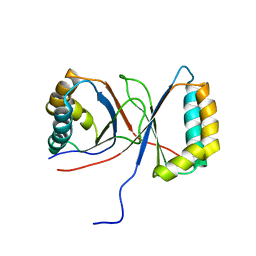 | |
7L1F
 
 | | SARS-CoV-2 RdRp in complex with 4 Remdesivir monophosphate | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (5'-R(P*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*UP*CP*UP*UP*AP*G)-3'), ... | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2020-12-14 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication.
Mol.Cell, 81, 2021
|
|
7LMQ
 
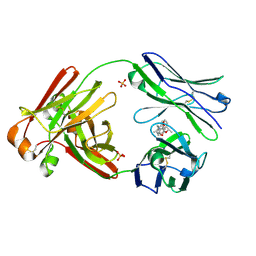 | |
7LMO
 
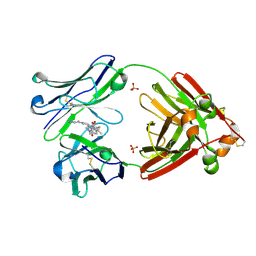 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 34 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-7-(1H-imidazole-5-carbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione] | | Descriptor: | (5~{R})-3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-7-(1~{H}-imidazol-4-ylcarbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7LMN
 
 | |
7LMR
 
 | |
7LMP
 
 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 36 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-8-(1H-imidazole-4-carbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione] | | Descriptor: | 3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-8-(1~{H}-imidazol-5-ylcarbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
1TES
 
 | | OXYGEN BINDING MUSCLE PROTEIN | | Descriptor: | METHYLETHYLAMINE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Phillips Jr, G.N, Olson, J.S. | | Deposit date: | 1996-05-06 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of NO-induced oxidation of myoglobin and hemoglobin.
Biochemistry, 35, 1996
|
|
6ENK
 
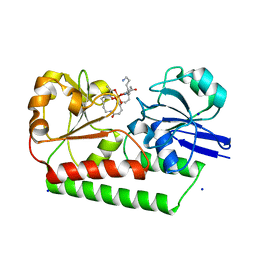 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
2I9Y
 
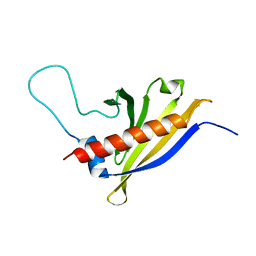 | | Solution structure of Arabidopsis thaliana protein At1g70830, a member of the major latex protein family | | Descriptor: | major latex protein-like protein 28 or MLP-like protein 28 | | Authors: | Volkman, B.F, de la Cruz, N.B, Lytle, B.L, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-09-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structures of two Arabidopsis thaliana major latex proteins represent novel helix-grip folds.
Proteins, 76, 2009
|
|
2IVY
 
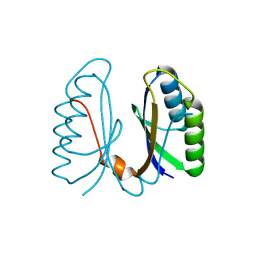 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
1VJH
 
 | | Crystal structure of gene product of At1g24000 from Arabidopsis thaliana | | Descriptor: | Bet v I allergen family | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1H, 15N and 13C resonance assignments of the putative Bet v 1 family protein At1g24000.1 from Arabidopsis thaliana.
J.Biomol.Nmr, 32, 2005
|
|
1VK0
 
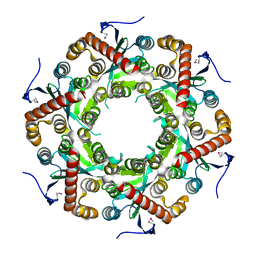 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450 | | Descriptor: | hypothetical protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-12 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450
To be published
|
|
1ZWJ
 
 | | X-ray structure of galt-like protein from arabidopsis thaliana AT5G18200 | | Descriptor: | ZINC ION, putative galactose-1-phosphate uridyl transferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, McCoy, J.G, Johnson, K.A, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of an ADP-Glucose Phosphorylase from
Arabidopsis thaliana
Biochemistry, 45, 2006
|
|
2Q3O
 
 | | Ensemble refinement of the protein crystal structure of 12-oxo-phytodienoate reductase isoform 3 | | Descriptor: | 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q3M
 
 | | Ensemble refinement of the protein crystal structure of an Arabidopsis thaliana putative steroid sulphotransferase | | Descriptor: | Flavonol sulfotransferase-like, MALONIC ACID | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q3P
 
 | | Ensemble refinement of the protein crystal structure of At3g17210 from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, Uncharacterized protein At3g17210 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2VV5
 
 | | The open structure of MscS | | Descriptor: | SMALL-CONDUCTANCE MECHANOSENSITIVE CHANNEL | | Authors: | Wang, W, Dong, C, Johnson, K.A, Naismith, J.H. | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Structure of an Open Form of an E. Coli Mechanosensitive Channel at 3.45 A Resolution.
Science, 321, 2008
|
|
2WR8
 
 | | Structure of Pyrococcus horikoshii SAM hydroxide adenosyltransferase in complex with SAH | | Descriptor: | PUTATIVE UNCHARACTERIZED PROTEIN PH0463, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMahon, S.A, Deng, H, O'Hagan, D, Johnson, K.A, Naismith, J.H. | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Mechanistic Insights Into Water Activation in Sam Hydroxide Adenosyltransferase (Duf-62).
Chembiochem, 10, 2009
|
|
2JC2
 
 | | The crystal structure of the natural F112L human sorcin mutant | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Franceschini, S, Ilari, A, Colotti, G, Chiancone, E. | | Deposit date: | 2006-12-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Impaired Function of the Natural F112L Sorcin Mutant: X-Ray Crystal Structure, Calcium Affinity, and Interaction with Annexin Vii and the Ryanodine Receptor.
Faseb J., 22, 2008
|
|
