7FII
 
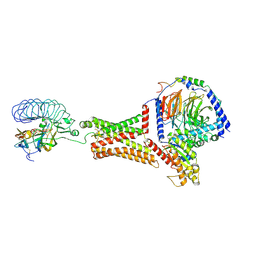 | | luteinizing hormone/choriogonadotropin receptor-chorionic gonadotropin-Gs complex | | Descriptor: | Choriogonadotropin subunit beta 3, Engineered Guanine nucleotide-binding protein G(s) subunit alpha, Glycoprotein hormones alpha chain, ... | | Authors: | Duan, J, Xu, P, Cheng, X, Mao, C, Croll, T, He, X, Shi, J, Luan, X, Yin, W, You, E, Liu, Q, Zhang, S, Jiang, H, Zhang, Y, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of full-length glycoprotein hormone receptor signalling complexes.
Nature, 598, 2021
|
|
7FIJ
 
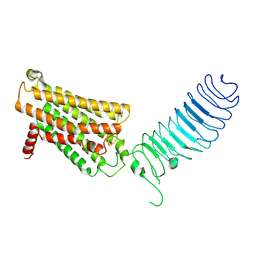 | | luteinizing hormone/choriogonadotropin receptor | | Descriptor: | Lutropin-choriogonadotropic hormone receptor | | Authors: | Duan, J, Xu, P, Cheng, X, Mao, C, Croll, T, He, X, Shi, J, Luan, X, Yin, W, You, E, Liu, Q, Zhang, S, Jiang, H, Zhang, Y, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of full-length glycoprotein hormone receptor signalling complexes.
Nature, 598, 2021
|
|
7FIG
 
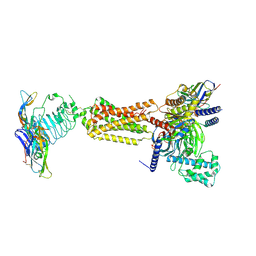 | | luteinizing hormone/choriogonadotropin receptor(S277I)-chorionic gonadotropin-Gs complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Camelid antibody VHH fragment Nb35, Choriogonadotropin subunit beta 3, ... | | Authors: | Duan, J, Xu, P, Cheng, X, Mao, C, Croll, T, He, X, Shi, J, Luan, X, Yin, W, You, E, Liu, Q, Zhang, S, Jiang, H, Zhang, Y, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of full-length glycoprotein hormone receptor signalling complexes.
Nature, 598, 2021
|
|
7FIH
 
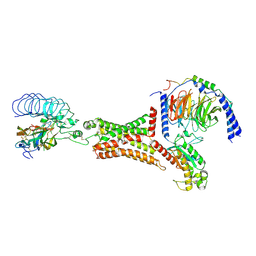 | | luteinizing hormone/choriogonadotropin receptor(S277I)-chorionic gonadotropin-Gs-Org43553 complex | | Descriptor: | 5-azanyl-N-tert-butyl-2-methylsulfanyl-4-[3-(2-morpholin-4-ylethanoylamino)phenyl]thieno[2,3-d]pyrimidine-6-carboxamide, Choriogonadotropin subunit beta 3, Engineered Guanine nucleotide-binding protein G(s) subunit alpha, ... | | Authors: | Duan, J, Xu, P, Cheng, X, Mao, C, Croll, T, He, X, Shi, J, Luan, X, Yin, W, You, E, Liu, Q, Zhang, S, Jiang, H, Zhang, Y, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of full-length glycoprotein hormone receptor signalling complexes.
Nature, 598, 2021
|
|
1SEK
 
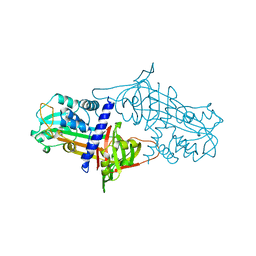 | | THE STRUCTURE OF ACTIVE SERPIN K FROM MANDUCA SEXTA AND A MODEL FOR SERPIN-PROTEASE COMPLEX FORMATION | | Descriptor: | SERPIN K | | Authors: | Li, J, Wang, Z, Canagarajah, B, Jiang, H, Kanost, M, Goldsmith, E.J. | | Deposit date: | 1998-03-06 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of active serpin 1K from Manduca sexta.
Structure Fold.Des., 7, 1999
|
|
5BTR
 
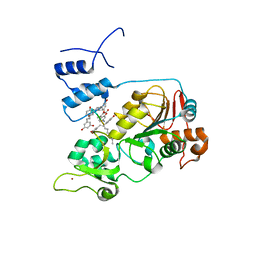 | | Crystal structure of SIRT1 in complex with resveratrol and an AMC-containing peptide | | Descriptor: | AMC-containing peptide, NAD-dependent protein deacetylase sirtuin-1, RESVERATROL, ... | | Authors: | Cao, D, Wang, M, Qiu, X, Liu, D, Jiang, H, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol
Genes Dev., 29, 2015
|
|
5BWL
 
 | |
2PVP
 
 | |
2PWX
 
 | | Crystal structure of G11A mutant of SARS-CoV 3C-like protease | | Descriptor: | 3C-like proteinase | | Authors: | Chen, S, Hu, T, Jiang, H, Shen, X. | | Deposit date: | 2007-05-14 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations.
J.Biol.Chem., 283, 2008
|
|
2QGH
 
 | | Crystal structure of diaminopimelate decarboxylase from Helicobacter pylori complexed with L-lysine | | Descriptor: | Diaminopimelate decarboxylase, GLYCEROL, LYSINE, ... | | Authors: | Hu, T, Wu, D, Jiang, H, Shen, X. | | Deposit date: | 2007-06-28 | | Release date: | 2008-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diaminopimelate decarboxylase from Helicobacter pylori complexed with L-lysine
TO BE PUBLISHED
|
|
2RJG
 
 | | Crystal structure of biosynthetic alaine racemase from Escherichia coli | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
2RJH
 
 | | Crystal structure of biosynthetic alaine racemase in D-cycloserine-bound form from Escherichia coli | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
7WKD
 
 | | TRH-TRHR G protein complex | | Descriptor: | Gq, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Y, Cai, H, You, C, He, X, Yuan, Q, Jiang, H, Cheng, X, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-01-09 | | Release date: | 2022-06-15 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into ligand binding and activation of the human thyrotropin-releasing hormone receptor.
Cell Res., 32, 2022
|
|
3B8V
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221K | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8W
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221P | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8T
 
 | | Crystal structure of Escherichia coli alaine racemase mutant P219A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8U
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3C5Q
 
 | | Crystal structure of diaminopimelate decarboxylase (I148L mutant) from Helicobacter pylori complexed with L-lysine | | Descriptor: | Diaminopimelate decarboxylase, GLYCEROL, LYSINE, ... | | Authors: | Hu, T, Wu, D, Jiang, H, Shen, X. | | Deposit date: | 2008-02-01 | | Release date: | 2008-05-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of diaminopimelate decarboxylase from Helicobacter pylori
To be Published
|
|
3CVA
 
 | | Human Bcl-xL containing a Trp to Ala mutation at position 137 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | Feng, Y, Zhang, L, Hu, T, Shen, X, Chen, K, Jiang, H, Liu, D. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved hydrophobic core at Bcl-x(L) mediates its structural stability and binding affinity with BH3-domain peptide of pro-apoptotic protein
Arch.Biochem.Biophys., 484, 2009
|
|
3DOO
 
 | | Crystal structure of shikimate dehydrogenase from Staphylococcus epidermidis complexed with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | Authors: | Han, C, Hu, T, Wu, D, Zhou, J, Shen, X, Qu, D, Jiang, H. | | Deposit date: | 2008-07-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Febs J., 276, 2009
|
|
3DON
 
 | | Crystal structure of shikimate dehydrogenase from Staphylococcus epidermidis | | Descriptor: | GLYCEROL, Shikimate dehydrogenase | | Authors: | Han, C, Hu, T, Wu, D, Zhou, J, Shen, X, Qu, D, Jiang, H. | | Deposit date: | 2008-07-05 | | Release date: | 2009-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Febs J., 276, 2009
|
|
3F9G
 
 | |
3F9F
 
 | |
3F9H
 
 | |
3F9E
 
 | |
