3US6
 
 | | Crystal Structure of Histidine-containing Phosphotransfer Protein MtHPt1 from Medicago truncatula | | Descriptor: | Histidine-containing Phosphotransfer Protein type 1, MtHPt1 | | Authors: | Ruszkowski, M, Brzezinski, K, Jedrzejczak, R, Dauter, M, Dauter, Z, Sikorski, M, Jaskolski, M. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Medicago truncatula histidine-containing phosphotransfer protein: Structural and biochemical insights into the cytokinin transduction pathway in plants.
Febs J., 280, 2013
|
|
6WLC
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WXC
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6X4I
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with 3'-uridinemonophosphate | | Descriptor: | 1,2-ETHANEDIOL, 3'-URIDINEMONOPHOSPHATE, SODIUM ION, ... | | Authors: | Chang, C, Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6X1B
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-R(*GP*U)-3'), PHOSPHATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-18 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
5JYB
 
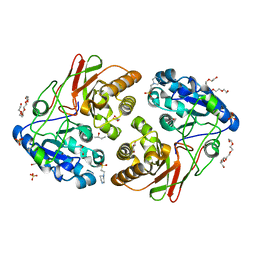 | | Crystal structure of 3 mutant of Ba3275 (S116A, E243A, H313A), the member of S66 family of serine peptidases | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nocek, B, Jedrzejczak, R, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of 3 mutant of Ba3275 (S116A, E243A, H313A), the member of S66 family of serine peptidases
To Be Published
|
|
5KCK
 
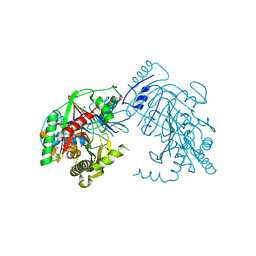 | | Crystal structure of anthranilate synthase component I from Streptococcus pneumoniae TIGR4 | | Descriptor: | Anthranilate synthase component I, GLYCEROL | | Authors: | Chang, C, Michalska, K, Bigelow, L, Jedrzejczak, R, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-06 | | Release date: | 2016-06-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of anthranilate synthase component I from Streptococcus pneumoniae TIGR4
To Be Published
|
|
5KBP
 
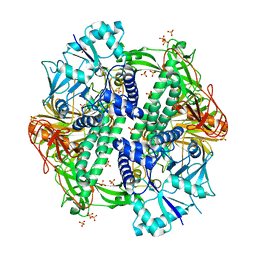 | | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583 | | Descriptor: | Glycosyl hydrolase, family 38, SULFATE ION | | Authors: | Tan, K, Chhor, G, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583
To Be Published
|
|
5L07
 
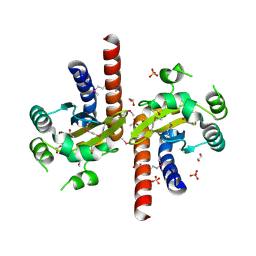 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Quorum-sensing transcriptional activator, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
5L09
 
 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica in complex with 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide, ACETIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
5L10
 
 | | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winan, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-07-28 | | Release date: | 2016-10-26 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia.
To Be Published
|
|
5T86
 
 | | Crystal structure of CDI complex from E. coli A0 34/86 | | Descriptor: | ACETATE ION, CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CDI complex from E. coli A0 34/86
To Be Published
|
|
3DO8
 
 | |
3GKU
 
 | | Crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940 | | Descriptor: | Probable RNA-binding protein | | Authors: | Tan, K, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940
To be Published
|
|
3H1Q
 
 | | Crystal structure of ethanolamine utilization protein EutJ from Carboxydothermus hydrogenoformans | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ethanolamine utilization protein EutJ | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Kinney, J, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-13 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutJ from Carboxydothermus hydrogenoformans
To be Published
|
|
7RLR
 
 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7RL8
 
 | | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7RBS
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with human ISG15 | | Descriptor: | Papain-like protease, Ubiquitin-like protein ISG15, ZINC ION | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
3H36
 
 | | Structure of an uncharacterized domain in polyribonucleotide nucleotidyltransferase from Streptococcus mutans UA159 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Polyribonucleotide nucleotidyltransferase | | Authors: | Cuff, M.E, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an uncharacterized domain in polyribonucleotide nucleotidyltransferase from Streptococcus mutans UA159
TO BE PUBLISHED
|
|
3HDT
 
 | | Crystal structure of putative kinase from Clostridium symbiosum ATCC 14940 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, putative kinase | | Authors: | Chang, C, Freeman, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-05-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of putative kinase from Clostridium symbiosum ATCC 14940
To be Published
|
|
7S6P
 
 | | The crystal structure of human ISG15 | | Descriptor: | Ubiquitin-like protein ISG15 | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
6XKM
 
 | | Room Temperature Structure of SARS-CoV-2 NSP10/NSP16 Methyltransferase in a Complex with SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3IC3
 
 | | Structure of a putative pyruvate dehydrogenase from the photosynthetic bacterium Rhodopseudomonas palustrus CGA009 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Cuff, M.E, Tesar, C, Jedrzejczak, R, Mckinlay, J.B, Harwood, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-09-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a putative pyruvate dehydrogenase from the photosynthetic bacterium Rhodopseudomonas palustrus CGA009
TO BE PUBLISHED
|
|
7THH
 
 | | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-11 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein
to be published
|
|
3IV3
 
 | | The Structure of a putative tagatose 1,6-aldolase from Streptococcus mutans | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Cuff, M.E, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of a putative tagatose 1,6-aldolase from Streptococcus mutans
TO BE PUBLISHED
|
|
