7ZEM
 
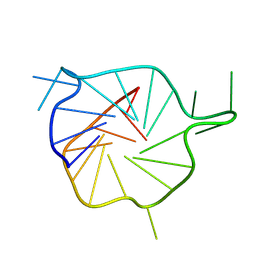 | |
7ZEO
 
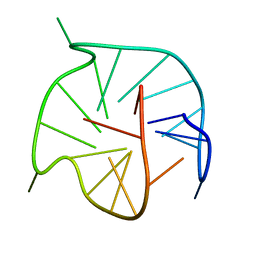 | |
7ZEK
 
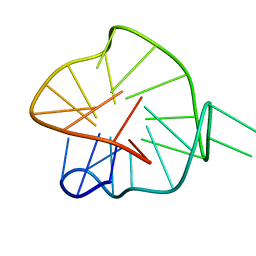 | |
8R4E
 
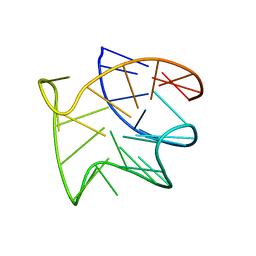 | |
8R4W
 
 | |
8RZX
 
 | |
8PSE
 
 | |
2MD3
 
 | |
2MD4
 
 | |
2MD2
 
 | |
2MD1
 
 | |
8PSI
 
 | |
8PSC
 
 | |
8PSB
 
 | | Three-layered parallel G-quadruplex with snapback loop from a G-rich sequence with five G-runs | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*AP*GP*GP*GP*CP*GP*GP*CP*GP*GP*GP*GP*AP*CP*GP*GP*GP*T)-3') | | Authors: | Vianney, Y.M, Schroeder, N, Jana, J, Chojetzki, G, Weisz, K. | | Deposit date: | 2023-07-13 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | Showcasing Different G-Quadruplex Folds of a G-Rich Sequence: Between Rule-Based Prediction and Butterfly Effect.
J.Am.Chem.Soc., 145, 2023
|
|
8S1W
 
 | |
4HH4
 
 | | Structure of the CcbJ Methyltransferase from Streptomyces caelestis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CcbJ, GLYCEROL, ... | | Authors: | Bauer, J.A, Ondrovicova, G, Kutejova, E, Janata, J. | | Deposit date: | 2012-10-09 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and possible mechanism of the CcbJ methyltransferase from Streptomyces caelestis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HGZ
 
 | | Structure of the CcbJ Methyltransferase from Streptomyces caelestis | | Descriptor: | 1,2-ETHANEDIOL, CcbJ, LITHIUM ION, ... | | Authors: | Bauer, J.A, Ondrovicova, G, Kutejova, E, Janata, J. | | Deposit date: | 2012-10-09 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and possible mechanism of the CcbJ methyltransferase from Streptomyces caelestis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HGY
 
 | | Structure of the CcbJ Methyltransferase from Streptomyces caelestis | | Descriptor: | 1,2-ETHANEDIOL, CcbJ, SULFATE ION | | Authors: | Bauer, J.A, Ondrovicova, G, Kutejova, E, Janata, J. | | Deposit date: | 2012-10-09 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and possible mechanism of the CcbJ methyltransferase from Streptomyces caelestis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6Q9N
 
 | | Crystal structure of PBP2a from MRSA in complex with piperacillin and quinazolinone | | Descriptor: | 3-[2-[(~{E})-2-(4-ethynylphenyl)ethenyl]-4-oxidanylidene-quinazolin-3-yl]benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Martinez-Caballero, S, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Quinazolinone Allosteric Inhibitor of PBP 2a Synergizes with Piperacillin and Tazobactam against Methicillin-Resistant Staphylococcus aureus.
Antimicrob.Agents Chemother., 63, 2019
|
|
6H5O
 
 | |
2M2C
 
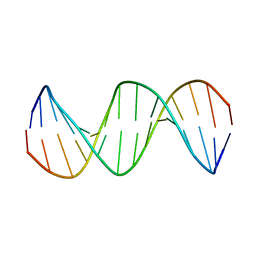 | | Solution structure of Duplex DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*AP*GP*CP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*AP*TP*GP*CP*TP*AP*CP*GP*CP*G)-3') | | Authors: | Ghosh, A, Kar, R.K, Chatterjee, S, Bhunia, A. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Indolicidin targets duplex DNA: structural and mechanistic insight through a combination of spectroscopy and microscopy.
Chemmedchem, 9, 2014
|
|
7ELJ
 
 | |
7O1H
 
 | |
8RW2
 
 | |
5XEP
 
 | | Crystal structure of BRP39, a chitinase-like protein, at 2.6 Angstorm resolution | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-3-like protein 1 | | Authors: | Mohanty, A.K, Fisher, A.J, Choudhary, S, Kaushik, J.K. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of BRP39, a signalling glycoprotein expressed during mammary gland apoptosis.
To be published
|
|
