1J9L
 
 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH VANADATE | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | 著者 | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | 登録日 | 2001-05-28 | | 公開日 | 2001-09-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1OHG
 
 | | STRUCTURE OF THE DSDNA BACTERIOPHAGE HK97 MATURE EMPTY CAPSID | | 分子名称: | CHLORIDE ION, MAJOR CAPSID PROTEIN, SULFATE ION | | 著者 | Helgstrand, C, Wikoff, W.R, Duda, R.L, Hendrix, R.W, Johnson, J.E, Liljas, L. | | 登録日 | 2003-05-26 | | 公開日 | 2003-12-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | The Refined Structure of a Protein Catenane: The Hk97 Bacteriophage Capsid at 3.44A Resolution
J.Mol.Biol., 334, 2003
|
|
1PPT
 
 | | X-RAY ANALYSIS (1.4-ANGSTROMS RESOLUTION) OF AVIAN PANCREATIC POLYPEPTIDE. SMALL GLOBULAR PROTEIN HORMONE | | 分子名称: | AVIAN PANCREATIC POLYPEPTIDE, ZINC ION | | 著者 | Blundell, T.L, Pitts, J.E, Tickle, I.J, Wood, S.P. | | 登録日 | 1981-01-16 | | 公開日 | 1981-02-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | X-ray analysis (1. 4-A resolution) of avian pancreatic polypeptide: Small globular protein hormone.
Proc.Natl.Acad.Sci.Usa, 78, 1981
|
|
3Q51
 
 | |
3Q50
 
 | |
1QJF
 
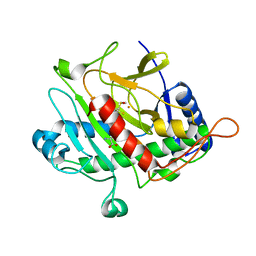 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (Monocyclic Sulfoxide - Fe COMPLEX) | | 分子名称: | 1-[(1S)-CARBOXY-2-(METHYLSULFINYL)ETHYL]-(3R)-[(5S)-5-AMINO-5-CARBOXYPENTANAMIDO]-(4R)-SULFANYLAZETIDIN-2-ONE, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | 著者 | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | 登録日 | 1999-06-23 | | 公開日 | 2000-06-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
1QJE
 
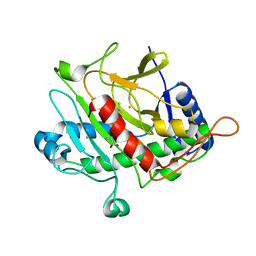 | | Isopenicillin N synthase from Aspergillus nidulans (IP1 - Fe complex) | | 分子名称: | FE (II) ION, ISOPENICILLIN N, ISOPENICILLIN N SYNTHASE, ... | | 著者 | Burzlaff, N.I, Clifton, I.J, Rutledge, P.J, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | 登録日 | 1999-06-23 | | 公開日 | 2000-06-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
1QIQ
 
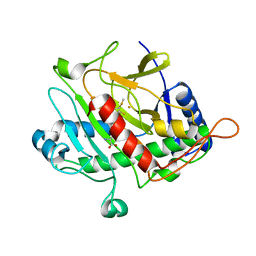 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACmC Fe COMPLEX) | | 分子名称: | FE (III) ION, ISOPENICILLIN N SYNTHASE, N-[N-[2-AMINO-6-OXO-HEXANOIC ACID-6-YL]CYSTEINYL]-S-METHYLCYSTEINE, ... | | 著者 | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | 登録日 | 1999-06-15 | | 公開日 | 2000-06-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction.
Nature, 401, 1999
|
|
1A5J
 
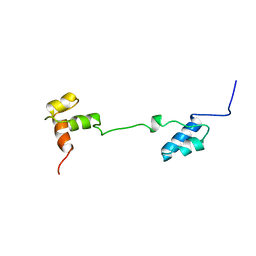 | | CHICKEN B-MYB DNA BINDING DOMAIN, REPEAT 2 AND REPEAT3, NMR, 32 STRUCTURES | | 分子名称: | B-MYB | | 著者 | Mcintosh, P.B, Carr, M.D, Wollborn, U, Frenkiel, T.A, Feeney, J, Mccormick, J.E, Klempnauer, K.H. | | 登録日 | 1998-02-16 | | 公開日 | 1998-07-01 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the B-Myb DNA-binding domain: a possible role for conformational instability of the protein in DNA binding and control of gene expression.
Biochemistry, 37, 1998
|
|
1ANJ
 
 | | ALKALINE PHOSPHATASE (K328H) | | 分子名称: | ALKALINE PHOSPHATASE, PHOSPHATE ION, ZINC ION | | 著者 | Murphy, J.E, Tibbitts, T.T, Kantrowitz, E.R. | | 登録日 | 1995-09-06 | | 公開日 | 1996-01-29 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mutations at positions 153 and 328 in Escherichia coli alkaline phosphatase provide insight towards the structure and function of mammalian and yeast alkaline phosphatases.
J.Mol.Biol., 253, 1995
|
|
1BGF
 
 | |
1BRZ
 
 | | SOLUTION STRUCTURE OF THE SWEET PROTEIN BRAZZEIN, NMR, 43 STRUCTURES | | 分子名称: | BRAZZEIN | | 著者 | Caldwell, J.E, Abildgaard, F, Dzakula, Z, Ming, D, Hellekant, G, Markley, J.L. | | 登録日 | 1998-03-12 | | 公開日 | 1998-07-01 | | 最終更新日 | 2019-12-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the thermostable sweet-tasting protein brazzein.
Nat.Struct.Biol., 5, 1998
|
|
1CWP
 
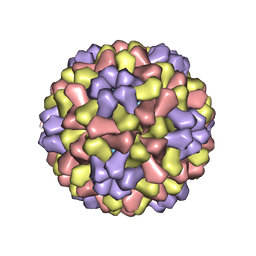 | | STRUCTURES OF THE NATIVE AND SWOLLEN FORMS OF COWPEA CHLOROTIC MOTTLE VIRUS DETERMINED BY X-RAY CRYSTALLOGRAPHY AND CRYO-ELECTRON MICROSCOPY | | 分子名称: | Coat protein, RNA (5'-R(*AP*U)-3'), RNA (5'-R(*AP*UP*AP*U)-3') | | 著者 | Speir, J.A, Johnson, J.E, Munshi, S, Wang, G, Timothy, S, Baker, T.S. | | 登録日 | 1995-05-22 | | 公開日 | 1995-05-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy.
Structure, 3, 1995
|
|
1CLD
 
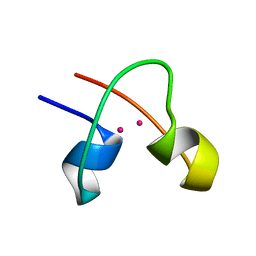 | | DNA-binding protein | | 分子名称: | CADMIUM ION, CD2-LAC9 | | 著者 | Gardner, K.H, Coleman, J.E. | | 登録日 | 1995-06-06 | | 公開日 | 1995-09-15 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Kluyveromyces lactis LAC9 Cd2 Cys6 DNA-binding domain.
Nat.Struct.Biol., 2, 1995
|
|
1DSU
 
 | |
5W8W
 
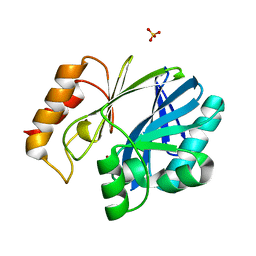 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 - new refinement | | 分子名称: | Metallo-beta-lactamase type 2, SULFATE ION, UNKNOWN ATOM OR ION, ... | | 著者 | Gonzalez, J.M, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Wlodawer, A, Gonzalez, M.M, Vila, A.J. | | 登録日 | 2017-06-22 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5WEQ
 
 | |
5WHV
 
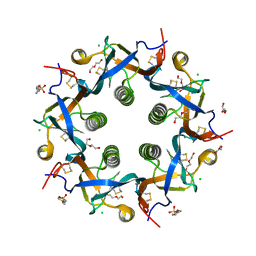 | | Crystal structure of ArtB | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ArtB protein, CALCIUM ION, ... | | 著者 | Gao, X, Galan, J.E. | | 登録日 | 2017-07-18 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Evolution of host adaptation in the Salmonella typhoid toxin.
Nat Microbiol, 2, 2017
|
|
5Y8R
 
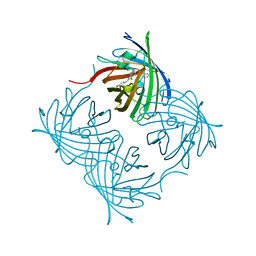 | | ZsYellow at pH 3.5 | | 分子名称: | GFP-like fluorescent chromoprotein FP538 | | 著者 | Bae, J.E, Kim, I.J, Nam, K.H. | | 登録日 | 2017-08-21 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y8Q
 
 | | ZsYellow at pH 8.0 | | 分子名称: | GFP-like fluorescent chromoprotein FP538 | | 著者 | Bae, J.E, Kim, I.J, Nam, K.H. | | 登録日 | 2017-08-21 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y4J
 
 | |
5Y4I
 
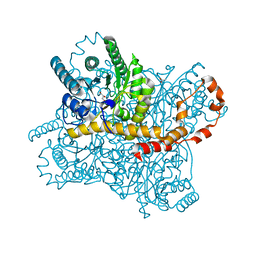 | | Crystal structure of glucose isomerase in complex with glycerol in one metal binding mode | | 分子名称: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Bae, J.E, Kim, I.J, Nam, K.H. | | 登録日 | 2017-08-03 | | 公開日 | 2017-09-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Crystal structure of glucose isomerase in complex with xylitol inhibitor in one metal binding mode
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Z6V
 
 | |
6APX
 
 | | Crystal structure of human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion in complex with the monobody YSX1 | | 分子名称: | GLYCEROL, Maltose-binding periplasmic protein,Dual specificity protein phosphatase 1, Monobody YSX1, ... | | 著者 | Gumpena, R, Lountos, G.T, Sreejith, R.K, Tropea, J.E, Cherry, S, Waugh, D.S. | | 登録日 | 2017-08-18 | | 公開日 | 2017-11-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.491 Å) | | 主引用文献 | Crystal structure of the human dual specificity phosphatase 1 catalytic domain.
Protein Sci., 27, 2018
|
|
6ARK
 
 | | Crystal Structure of compound 10 covalently bound to K-Ras G12C | | 分子名称: | (3R)-N-(6-bromonaphthalen-2-yl)-3-hydroxy-1-propanoyl-L-prolinamide, GLYCEROL, GTPase KRas, ... | | 著者 | Nnadi, C.I, Jenkins, M.L, Gentile, D.R, Bateman, L.A, Zaidman, D, Balius, T.E, Nomura, D.K, Burke, J.E, Shokat, K.M, London, N. | | 登録日 | 2017-08-22 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Novel K-Ras G12C Switch-II Covalent Binders Destabilize Ras and Accelerate Nucleotide Exchange.
J Chem Inf Model, 58, 2018
|
|
