2ZJ0
 
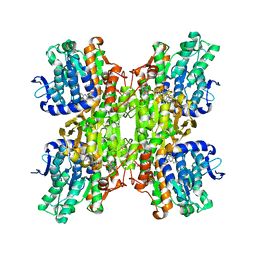 | | Crystal structure of Mycobacterium tuberculosis S-Adenosyl-L-homocysteine hydrolase in ternary complex with NAD and 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
2ZEC
 
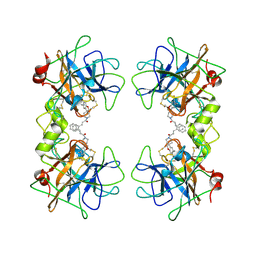 | | Potent, Nonpeptide Inhibitors of Human Mast Cell Tryptase | | Descriptor: | 1-[1'-(3-phenylacryloyl)spiro[1-benzofuran-3,4'-piperidin]-5-yl]methanamine, Tryptase beta 2 | | Authors: | Spurlino, J.C, Lewandowski, F, Milligan, C. | | Deposit date: | 2007-12-08 | | Release date: | 2008-12-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2Z9I
 
 | |
2ZEB
 
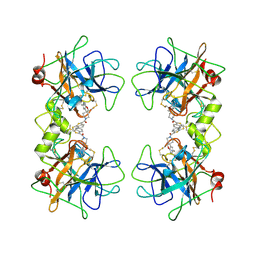 | | Potent, Nonpeptide Inhibitors of Human Mast Cell Tryptase | | Descriptor: | 1-(1'-{[3-(methylsulfanyl)-2-benzothiophen-1-yl]carbonyl}spiro[1-benzofuran-3,4'-piperidin]-5-yl)methanamine, Tryptase beta 2 | | Authors: | Spurlino, J.C, Lewandowski, F, Milligan, C. | | Deposit date: | 2007-12-08 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2ZA5
 
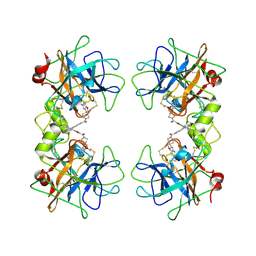 | | Crystal Structure of human tryptase with potent non-peptide inhibitor | | Descriptor: | (5-(aminomethyl)-2H-spiro[benzofuran-3,4'-piperidine]-1'-yl)(5-(phenylethynyl)furan-2-yl)methanone, Tryptase beta 2 | | Authors: | Spurlino, J.C, Barnakov, S.A, Lewandowski, F, Milligan, C. | | Deposit date: | 2007-10-02 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2ZIZ
 
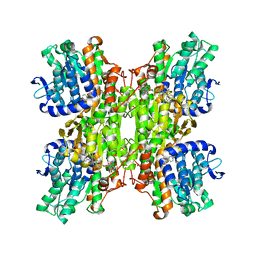 | | Crystal structure of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with NAD and 3-deazaadenosine | | Descriptor: | 3-DEAZA-ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
8ARL
 
 | | Plasmodium vivax PVP01_0000100 TRAg domain | | Descriptor: | Tryptophan-rich antigen | | Authors: | Kundu, P, Deane, J.E, Rayner, J.C. | | Deposit date: | 2022-08-17 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a Plasmodium vivax Tryptophan Rich Antigen domain suggests a lipid binding function for a pan-Plasmodium multi-gene family.
Nat Commun, 14, 2023
|
|
7PLM
 
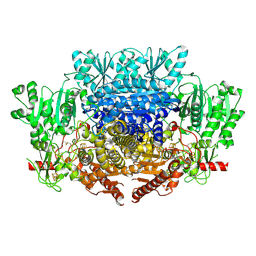 | | CryoEM reconstruction of pyruvate ferredoxin oxidoreductase (PFOR) in anaerobic conditions | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cherrier, M.V, Vernede, X, Fenel, D, Martin, L, Arragain, B, Neumann, E, Fontecilla Camps, J.C, Schoehn, G, Nicolet, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Oxygen-Sensitive Metalloprotein Structure Determination by Cryo-Electron Microscopy.
Biomolecules, 12, 2022
|
|
8BNB
 
 | |
8BOJ
 
 | |
8BP5
 
 | |
8BP4
 
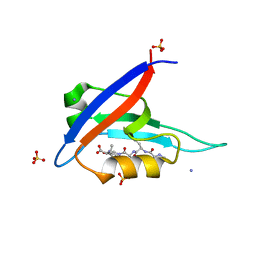 | |
8BPM
 
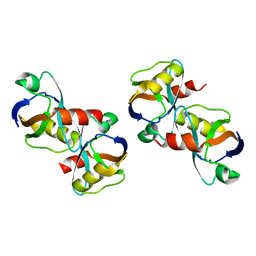 | |
1AA6
 
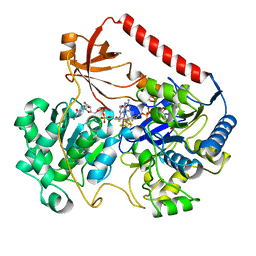 | | REDUCED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
2FRV
 
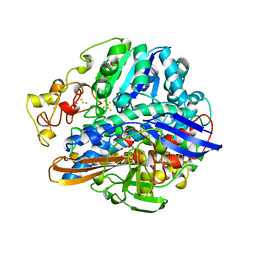 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF NI-FE HYDROGENASE | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1997-06-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of the [Nife] Hydrogenase Active Site: Evidence for Biologically Uncommon Fe Ligands
J.Am.Chem.Soc., 118, 1996
|
|
8BTY
 
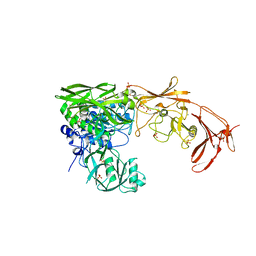 | | Structure of the active form of ScpB, the C5a-peptidase from Streptococcus agalactiae. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Kagawa, T.F, Cooney, J.C, Miclot, T, Cullen, R. | | Deposit date: | 2022-11-30 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 angstrom crystal structure of the C5a peptidase from Streptococcus agalactiae (ScpB) reveals an active site competent for catalysis.
Proteins, 92, 2024
|
|
8BQ8
 
 | |
8C9N
 
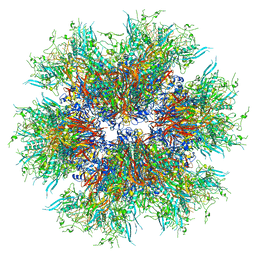 | | MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle | | Descriptor: | Penton protein | | Authors: | Bufton, J.C, Capin, J, Boruku, U, Garzoni, F, Schaffitzel, C, Berger, I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | In vitro generated antibodies guide thermostable ADDomer nanoparticle design for nasal vaccination and passive immunization against SARS-CoV-2.
Antib Ther, 6, 2023
|
|
8BUW
 
 | |
8BV7
 
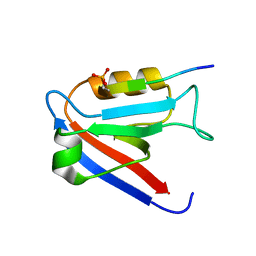 | |
8C1T
 
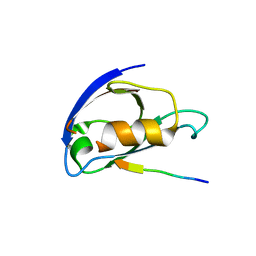 | |
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
8CTQ
 
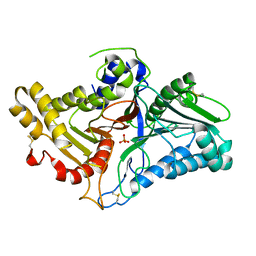 | | Crystal structure of engineered phospholipase D mutant superPLD 2-48 | | Descriptor: | PHOSPHATE ION, Phospholipase D | | Authors: | Tei, R, Bagde, S.R, Fromme, J.C, Baskin, J.M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Activity-based directed evolution of a membrane editor in mammalian cells.
Nat.Chem., 15, 2023
|
|
8CTP
 
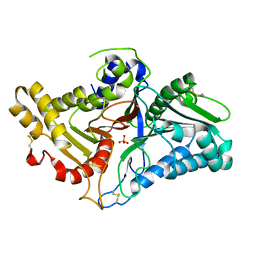 | | Crystal structure of engineered phospholipase D mutant superPLD 2-23 | | Descriptor: | PHOSPHATE ION, Phospholipase D | | Authors: | Tei, R, Bagde, S.R, Fromme, J.C, Baskin, J.M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-based directed evolution of a membrane editor in mammalian cells.
Nat.Chem., 15, 2023
|
|
8COY
 
 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) in complex with inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
