5D4H
 
 | | High-resolution nitrite complex of a copper nitrite reductase determined by synchrotron radiation crystallography | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4J
 
 | | Chloride-bound form of a copper nitrite reductase from Alcaligenes faecals | | Descriptor: | ACETIC ACID, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YSC
 
 | | Completely oxidized structure of copper nitrite reductase from Alcaligenes faecalis | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YSE
 
 | | High resolution synchrotron structure of copper nitrite reductase from Alcaligenes faecalis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5YC8
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS (Hg-derivative) | | Descriptor: | MERCURY (II) ION, Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZK8
 
 | | Crystal structure of M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKB
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with AF-DX 384 | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-[2-[(2S)-2-[(dipropylamino)methyl]piperidin-1-yl]ethyl]-6-oxidanylidene-5H-pyrido[2,3-b][1,4]benzodiazepine-11-carboxamide | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKC
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZK3
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with QNB | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2 | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
2CFQ
 
 | | Sugar Free Lactose Permease at neutral pH | | Descriptor: | LACTOSE PERMEASE, MERCURY (II) ION | | Authors: | Mirza, O, Guan, L, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Evidence for Induced Fit and a Mechanism for Sugar/H(+) Symport in Lacy.
Embo J., 25, 2006
|
|
2XUT
 
 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
2CFP
 
 | | Sugar Free Lactose Permease at acidic pH | | Descriptor: | LACTOSE PERMEASE, MERCURY (II) ION | | Authors: | Mirza, O, Guan, L, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Evidence for Induced Fit and a Mechanism for Sugar/H(+) Symport in Lacy.
Embo J., 25, 2006
|
|
5YFI
 
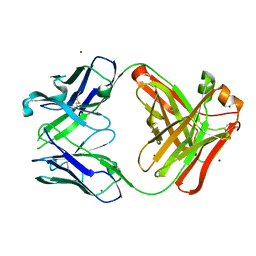 | | Crystal structure of the anti-human prostaglandin E receptor EP4 antibody Fab fragment | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, ZINC ION | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-21 | | Release date: | 2018-12-05 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YHL
 
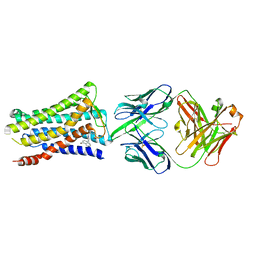 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and an antagonist Br-derivative | | Descriptor: | 4-[2-[[(2R)-2-(4-bromanylnaphthalen-1-yl)propanoyl]amino]-4-cyano-phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-28 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YWY
 
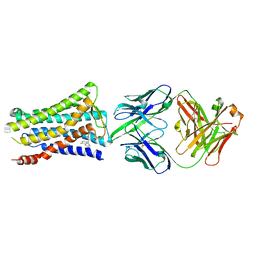 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208 | | Descriptor: | 4-[4-cyano-2-[[(2R)-2-(4-fluoranylnaphthalen-1-yl)propanoyl]amino]phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
3AOU
 
 | | Structure of the Na+ unbound rotor ring modified with N,N f-Dicyclohexylcarbodiimide of the Na+-transporting V-ATPase | | Descriptor: | DICYCLOHEXYLUREA, UNDECYL-MALTOSIDE, V-type sodium ATPase subunit K | | Authors: | Mizutani, K, Yamamoto, M, Yamato, I, Kakinuma, Y, Shirouzu, M, Yokoyama, S, Iwata, S, Murata, T. | | Deposit date: | 2010-10-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7ZBE
 
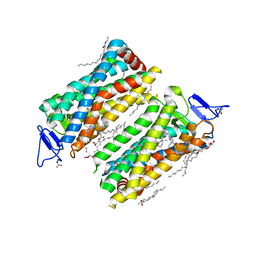 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SwissFEL) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
7ZBC
 
 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, S, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
7D7M
 
 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Nojima, S, Fujita, Y, Kimura, T.K, Nomura, N, Suno, R, Morimoto, K, Yamamoto, M, Noda, T, Iwata, S, Shigematsu, H, Kobayashi, T. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein.
Structure, 29, 2021
|
|
2V8N
 
 | | Wild-type Structure of Lactose Permease | | Descriptor: | LACTOSE PERMEASE | | Authors: | Guan, L, Mirza, O, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2007-08-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Determination of Wild-Type Lactose Permease.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2VYC
 
 | | Crystal Structure of Acid Induced Arginine Decarboxylase from E. coli | | Descriptor: | BIODEGRADATIVE ARGININE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Andrell, J, Hicks, M.G, Palmer, T, Carpenter, E.P, Iwata, S, Maher, M.J. | | Deposit date: | 2008-07-22 | | Release date: | 2009-03-31 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Acid Induced Arginine Decarboxylase from Escherichia Coli: Reversible Decamer Assembly Controls Enzyme Activity.
Biochemistry, 48, 2009
|
|
2VOC
 
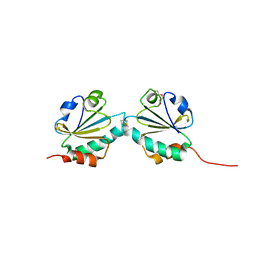 | | THIOREDOXIN A ACTIVE SITE MUTANTS FORM MIXED DISULFIDE DIMERS THAT RESEMBLE ENZYME-SUBSTRATE REACTION INTERMEDIATE | | Descriptor: | DI(HYDROXYETHYL)ETHER, THIOREDOXIN | | Authors: | Kouwen, T.R.H.M, Andrell, J, Schrijver, R, Dubois, J.Y.F, Maher, M.J, Iwata, S, Carpenter, E.P, van Dijl, J.M. | | Deposit date: | 2008-02-13 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thioredoxin A active-site mutants form mixed disulfide dimers that resemble enzyme-substrate reaction intermediates.
J. Mol. Biol., 379, 2008
|
|
2WDQ
 
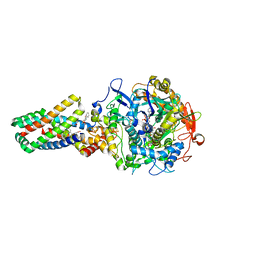 | | E. coli succinate:quinone oxidoreductase (SQR) with carboxin bound | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-03-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Escherichia coli succinate:quinone oxidoreductase with an occupied and empty quinone-binding site.
J. Biol. Chem., 284, 2009
|
|
2WDV
 
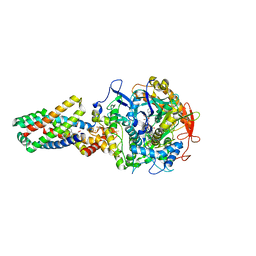 | | E. coli succinate:quinone oxidoreductase (SQR) with an empty quinone- binding pocket | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-03-26 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Escherichia Coli Succinate:Quinone Oxidoreductase with an Occupied and Empty Quinone- Binding Site.
J.Biol.Chem., 284, 2009
|
|
2WDR
 
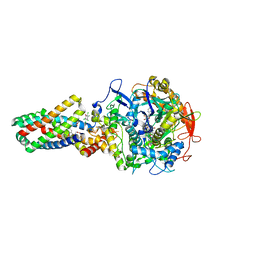 | | E. coli succinate:quinone oxidoreductase (SQR) with pentachlorophenol bound | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-03-25 | | Release date: | 2009-08-25 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Escherichia Coli Succinate:Quinone Oxidoreductase with an Occupied and Empty Quinone- Binding Site.
J.Biol.Chem., 284, 2009
|
|
