6JPP
 
 | |
8I5F
 
 | | Crystal structure of the DHR-2 domain of DOCK10 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 10 | | Authors: | Kukimoto-Niino, M, Mishima-Tsumagari, C, Fukui, Y, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2023-01-25 | | Release date: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
7DM9
 
 | |
7DMA
 
 | |
3A7K
 
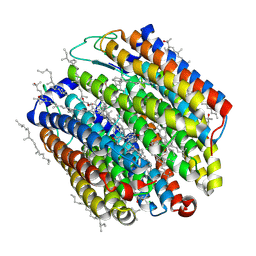 | | Crystal structure of halorhodopsin from Natronomonas pharaonis | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Kouyama, T. | | Deposit date: | 2009-09-27 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Light-Driven Chloride Pump Halorhodopsin from Natronomonas pharaonis.
J.Mol.Biol., 396, 2010
|
|
4FBZ
 
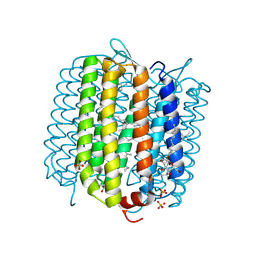 | | Crystal structure of deltarhodopsin from Haloterrigena thermotolerans | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, BACTERIORUBERIN, ... | | Authors: | Kouyama, T. | | Deposit date: | 2012-05-23 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of deltarhodopsin-3 from Haloterrigena thermotolerans
Proteins, 81, 2013
|
|
1MPG
 
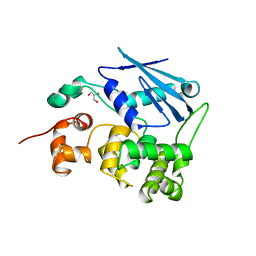 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | Authors: | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
3QBI
 
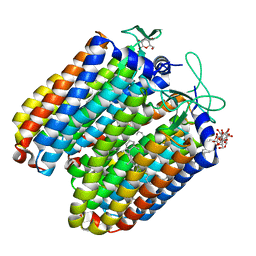 | |
3QBL
 
 | | Pharaonis halorhodopsin complexed with nitrate | | Descriptor: | BACTERIORUBERIN, Halorhodopsin, NITRATE ION, ... | | Authors: | Kouyama, T, Kanada, S. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of anion-free blue and yellow forms of pharaonis halorhodopsin
To be Published
|
|
3QBK
 
 | | Bromide-bound form of pharaonis halorhodopsin | | Descriptor: | BACTERIORUBERIN, BROMIDE ION, Halorhodopsin, ... | | Authors: | Kouyama, T, Kanada, S. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of an O-like blue form and an anion-free yellow form of pharaonis halorhodopsin
J.Mol.Biol., 413, 2011
|
|
3QBG
 
 | | Anion-free blue form of pharaonis halorhodopsin | | Descriptor: | BACTERIORUBERIN, Halorhodopsin, RETINAL, ... | | Authors: | Kouyama, T, Kanada, S. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of an O-like blue form and an anion-free yellow form of pharaonis halorhodopsin
J.Mol.Biol., 413, 2011
|
|
4FWG
 
 | | Crystal structure of the Lon-like protease MtaLonC in complex with lactacystin | | Descriptor: | Omuralide, open form, PHOSPHATE ION, ... | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FWD
 
 | | Crystal structure of the Lon-like protease MtaLonC in complex with bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-06-30 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FWH
 
 | | Crystal structure of the Lon-like protease MtaLonC in complex with MG262 | | Descriptor: | N-[(benzyloxy)carbonyl]-L-leucyl-N-[(1R)-1-(dihydroxyboranyl)-3-methylbutyl]-L-leucinamide, PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WXQ
 
 | | Serial femtosecond X-ray structure of human fatty acid-binding protein type-3 (FABP3) in complex with stearic acid (C18:0) determined using X-ray free-electron laser at SACLA | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Mizohata, E, Suzuki, M, Kakinouchi, K, Sugiyama, S, Murata, M, Sugahara, M, Nango, E, Tanaka, T, Tanaka, R, Tono, K, Song, C, Hatsui, T, Joti, Y, Yabashi, M, Iwata, S. | | Deposit date: | 2014-08-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3WUM
 
 | |
3WUL
 
 | |
3WXS
 
 | | Thaumatin structure determined by SPring-8 Angstrom Compact free electron Laser (SACLA) | | Descriptor: | L(+)-TARTARIC ACID, thaumatin I | | Authors: | Masuda, T, Nango, E, Sugahara, M, Mizohata, E, Tanaka, T, Tanaka, R, Suzuki, M, Mikami, B, Iwata, S. | | Deposit date: | 2014-08-07 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3WXT
 
 | |
3WXU
 
 | |
3VHZ
 
 | |
3VI0
 
 | |
3VVK
 
 | |
1VGP
 
 | |
1VGM
 
 | |
