8TRM
 
 | | Actin 1 from T. gondii in filaments bound to MgADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, MAGNESIUM ION | | Authors: | Hvorecny, K.L, Sladewski, T.E, Heaslip, A.T, Kollman, J.M. | | Deposit date: | 2023-08-09 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Toxoplasma gondii actin filaments are tuned for rapid disassembly and turnover.
Nat Commun, 15, 2024
|
|
8TRN
 
 | | Actin 1 from T. gondii in filaments bound to MgADP and jasplakinolide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, Jasplakinolide, ... | | Authors: | Hvorecny, K.L, Sladewski, T.E, Heaslip, A.T, Kollman, J.M. | | Deposit date: | 2023-08-09 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Toxoplasma gondii actin filaments are tuned for rapid disassembly and turnover.
Nat Commun, 15, 2024
|
|
5TNN
 
 | |
5TNK
 
 | |
5TNL
 
 | |
5TNG
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 14,15-EpETE hydrolysis intermediate | | Descriptor: | (5Z,8Z,11Z,14R,15R,17Z)-14,15-dihydroxyicosa-5,8,11,17-tetraenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNR
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 16,17-EpDPE hydrolysis intermediate | | Descriptor: | (4Z,7Z,10Z,13Z,16R,17R,19Z)-16,17-dihydroxydocosa-4,7,10,13,19-pentaenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNF
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 19,20-EpDPE hydrolysis intermediate | | Descriptor: | (4Z,7Z,10Z,13Z,16Z,19R,20R)-19,20-dihydroxydocosa-4,7,10,13,16-pentaenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNQ
 
 | |
5TNH
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 17,18-EpETE hydrolysis intermediate | | Descriptor: | (5Z,8Z,11Z,14Z,17R,18R)-17,18-dihydroxyicosa-5,8,11,14-tetraenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNS
 
 | |
5TNJ
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 4-Vinyl-1-cyclohexene 1,2-epoxide hydrolysis intermediate | | Descriptor: | (1R,2R,4R)-4-ethenylcyclohexane-1,2-diol, (1R,2R,4S)-4-ethenylcyclohexane-1,2-diol, (1S,2S,4R)-4-ethenylcyclohexane-1,2-diol, ... | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TND
 
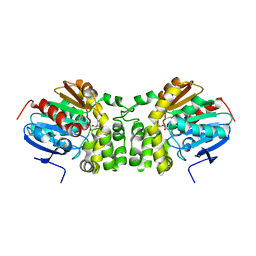 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 1,2-Epoxycyclohexane hydrolysis intermediate | | Descriptor: | (1R,2R)-cyclohexane-1,2-diol, (1S,2S)-cyclohexane-1,2-diol, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNM
 
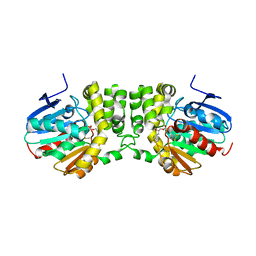 | |
5TNE
 
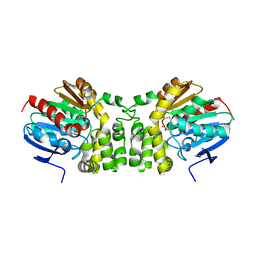 | |
5TNP
 
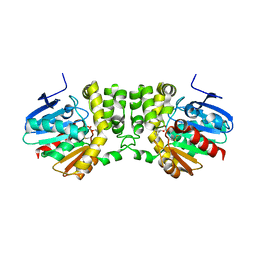 | |
5TNI
 
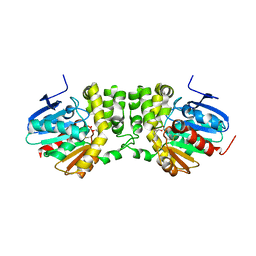 | |
8DBE
 
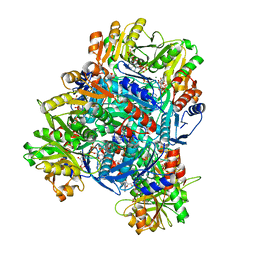 | | Human PRPS1 with ADP; Hexamer | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBO
 
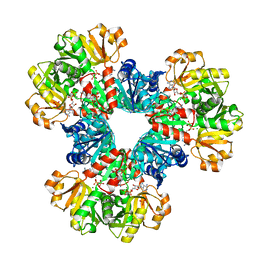 | |
8DBF
 
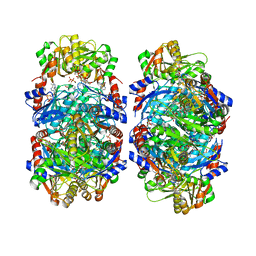 | | Human PRPS1 with ADP; Filament Interface | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBL
 
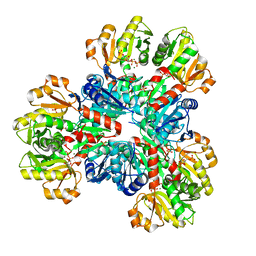 | | Human PRPS1 with Phosphate and PRPP; Hexamer | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBG
 
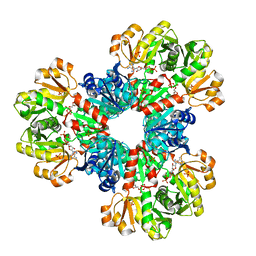 | | Human PRPS1 with Phosphate and ATP; Hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBN
 
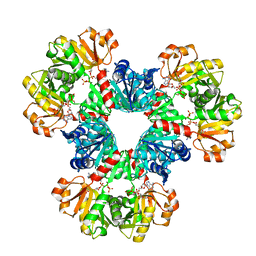 | | Human PRPS1-E307A engineered mutation with Phosphate, ATP, and R5P; Hexamer | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBJ
 
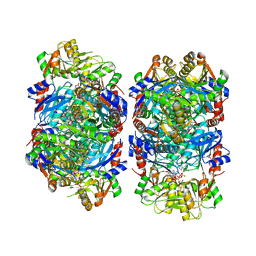 | | Human PRPS1 with Phosphate, ATP, and R5P; Filament Interface | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBK
 
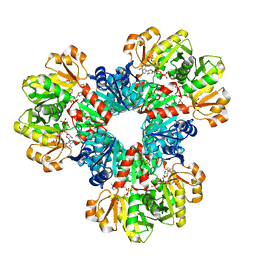 | | Human PRPS1 with Phosphate, ATP, and R5P; Hexamer with resolved catalytic loops | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
