8FX1
 
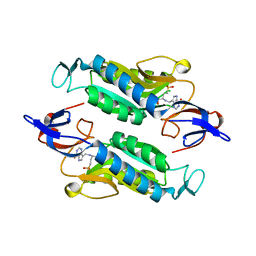 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (R)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FWY
 
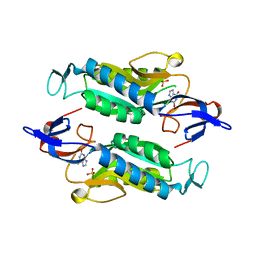 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to the dead-end complex xanthine and pyrophosphate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHATE ION, XANTHINE | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX3
 
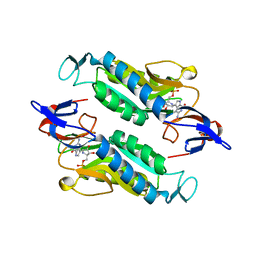 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-GP, showing the structure of the complete active site in its open conformation | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX0
 
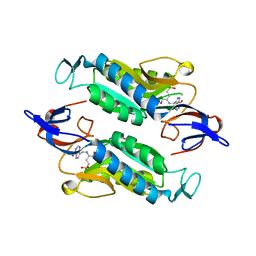 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (S)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX2
 
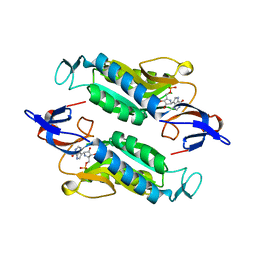 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-HP | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FWZ
 
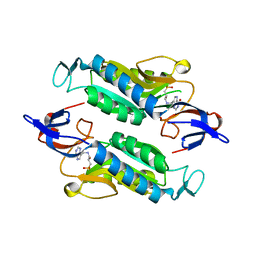 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Hydroxypropyl-Lin-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHATE ION, [(2R)-2-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}propyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
1OSH
 
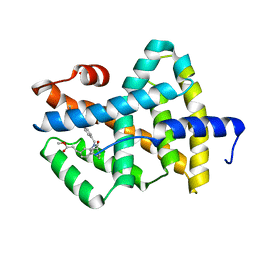 | | A Chemical, Genetic, and Structural Analysis of the nuclear bile acid receptor FXR | | Descriptor: | Bile acid receptor, METHYL 3-{3-[(CYCLOHEXYLCARBONYL){[4'-(DIMETHYLAMINO)BIPHENYL-4-YL]METHYL}AMINO]PHENYL}ACRYLATE | | Authors: | Downes, M, Verdecia, M.A, Roecker, A.J, Hughes, R, Hogenesch, J.B, Kast-Woelbern, H.R, Bowman, M.E, Ferrer, J.-L, Anisfeld, A.M, Edwards, P.A, Rosenfeld, J.M, Alvarez, J.G.A, Noel, J.P, Nicolaou, K.C, Evans, R.M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR
Mol.Cell, 11, 2003
|
|
6DNM
 
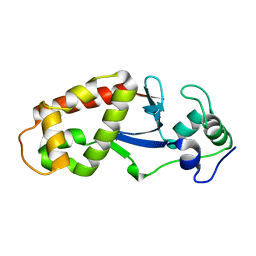 | |
6DRQ
 
 | |
6U7A
 
 | | Rv3722c in complex with kynurenine | | Descriptor: | (2S)-4-(2-aminophenyl)-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-hydroxyquinoline-2-carboxylic acid, ... | | Authors: | Mandyoli, L, Sacchettini, J. | | Deposit date: | 2019-09-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Aspartate aminotransferase Rv3722c governs aspartate-dependent nitrogen metabolism in Mycobacterium tuberculosis.
Nat Commun, 11, 2020
|
|
6U78
 
 | | Rv3722c in complex with glutamic acid | | Descriptor: | Aminotransferase, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Mandyoli, L, Sacchettini, J.C. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aspartate aminotransferase Rv3722c governs aspartate-dependent nitrogen metabolism in Mycobacterium tuberculosis.
Nat Commun, 11, 2020
|
|
8BE1
 
 | | SARS-CoV-2 RBD in complex with a Fab fragment of a neutralising antibody mRBD2 | | Descriptor: | Antibody heavy chain, Antibody light chain, SULFATE ION, ... | | Authors: | Lulla, A, Brear, P, Fischer, K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Microfluidics-enabled fluorescence-activated cell sorting of single pathogen-specific antibody secreting cells for the rapid discovery of monoclonal antibodies
Biorxiv, 2023
|
|
5VB3
 
 | | X-ray structure of nuclear receptor ROR-gammat Ligand Binding Domain + SRC2 peptide | | Descriptor: | Nuclear receptor ROR-gamma, SRC2 chimera, SODIUM ION | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
5VB7
 
 | | X-ray co-structure of nuclear receptor ROR-gammat Ligand Binding Domain with an agonist and SRC2 peptide | | Descriptor: | N-methyl-N'-(3-methylbut-2-en-1-yl)-N'-(3-phenoxyphenyl)-N-[trans-4-(pyridin-4-yl)cyclohexyl]urea, Nuclear receptor ROR-gamma, SRC2 chimera, ... | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
5VB6
 
 | |
5VB5
 
 | | X-ray co-structure of nuclear receptor ROR-gammat Ligand Binding Domain with an inverse agonist and SRC2 peptide | | Descriptor: | N-[(2R)-3-(4-{[3-(4-chlorophenyl)propanoyl]amino}phenyl)-1-(4-methylpiperidin-1-yl)-1-oxopropan-2-yl]-4-methylpentanamide, Nuclear receptor ROR-gamma, SRC2 chimera, ... | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
