8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8F3D
 
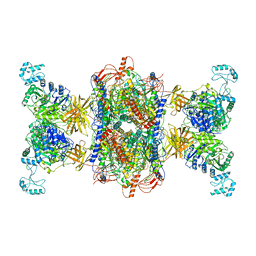 | | 3-methylcrotonyl-CoA carboxylase in filament, beta-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase alpha-subunit, 3-methylcrotonyl-CoA carboxylase beta-subunit, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
8F41
 
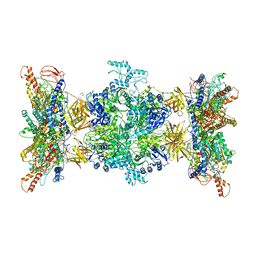 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
1NH9
 
 | | Crystal Structure of a DNA Binding Protein Mja10b from the hyperthermophile Methanococcus jannaschii | | Descriptor: | DNA-binding protein Alba | | Authors: | Wang, G, Bartlam, M, Guo, R, Yang, H, Xue, H, Liu, Y, Huang, L, Rao, Z. | | Deposit date: | 2002-12-19 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a DNA binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii
Protein Sci., 12, 2003
|
|
3BCC
 
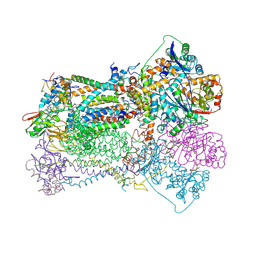 | | STIGMATELLIN AND ANTIMYCIN BOUND CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | ANTIMYCIN, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
5WE1
 
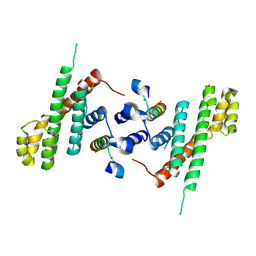 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | Protection of telomeres protein poz1,Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ZINC ION | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
5WE0
 
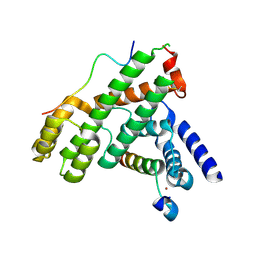 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
3F2W
 
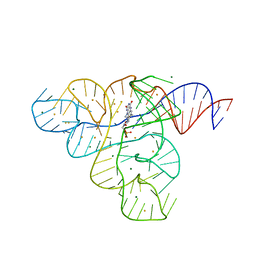 | |
3F4H
 
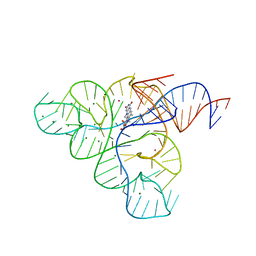 | | Crystal structure of the FMN riboswitch bound to roseoflavin | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, FMN riboswitch, MAGNESIUM ION, ... | | Authors: | Serganov, A.A, Huang, L. | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch.
Nature, 458, 2009
|
|
3F2Q
 
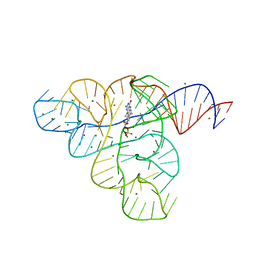 | |
3F4G
 
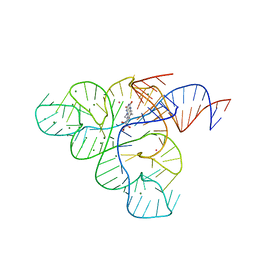 | |
3F2X
 
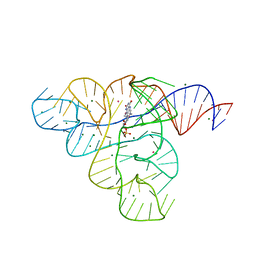 | |
3F2Y
 
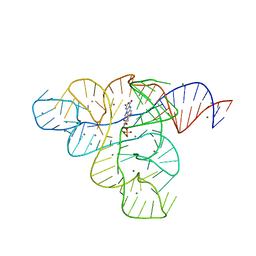 | |
3F30
 
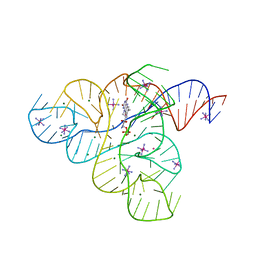 | |
3F2T
 
 | |
3F59
 
 | |
3F4E
 
 | |
4MM0
 
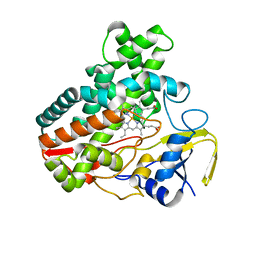 | | Crystal Structure Analysis of the Putative Thioether Synthase SgvP Involved in the Tailoring Step of Griseoviridin | | Descriptor: | P450-like monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhang, H, Huang, L, Yi, M, Cai, T, Zhang, H. | | Deposit date: | 2013-09-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of SgvP: a Putative Thioether Synthase Involved in the Tailoring Step of Griseoviridin
To be Published
|
|
8HB8
 
 | |
8HBA
 
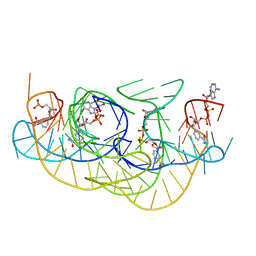 | |
8HB1
 
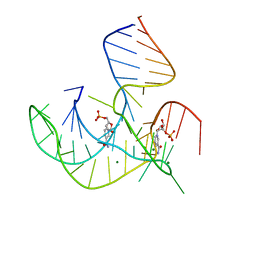 | | Crystal structure of NAD-II riboswitch (two strands) with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, RNA (30-MER), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
8HB3
 
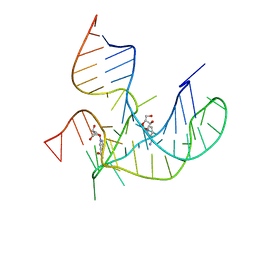 | | Crystal structure of NAD-II riboswitch (two strands) with NR | | Descriptor: | Nicotinamide riboside, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3') | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
