2M9G
 
 | |
6KLA
 
 | | Crystal structure of human c-KIT kinase domain in complex with compound 15a | | Descriptor: | Mast/stem cell growth factor receptor Kit, N-[6-(4-ethylpiperazin-1-yl)-2-methyl-pyrimidin-4-yl]-5-pyridin-4-yl-1,3-thiazol-2-amine | | Authors: | Wu, T.S, Peng, Y.H, Hsueh, C.C, Wu, S.Y. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Identification of a Multitargeted Tyrosine Kinase Inhibitor for the Treatment of Gastrointestinal Stromal Tumors and Acute Myeloid Leukemia.
J.Med.Chem., 62, 2019
|
|
4H00
 
 | | The crystal structure of mon-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
4H01
 
 | | The crystal structure of di-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
4GZ7
 
 | | The crystal structure of Apo-dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, dihydropyrimidinase | | Authors: | Hsien, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structure Requirements for Post-translational Modification
To be Published
|
|
4LCQ
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBI | | Descriptor: | (2S)-3-(carbamoylamino)-2-methylpropanoic acid, ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
4JBQ
 
 | | Novel Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules | | Descriptor: | Aurora Kinase A, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE | | Authors: | Wu, J.S, Leou, J.S, Peng, Y.H, Hsueh, C.C, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JBO
 
 | | Novel Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules | | Descriptor: | 1-(4-{2-[(6-{4-[2-(dimethylamino)ethoxy]phenyl}furo[2,3-d]pyrimidin-4-yl)amino]ethyl}phenyl)-3-phenylurea, Aurora kinase A | | Authors: | Wu, J.S, Leou, J.S, Peng, Y.H, Hsueh, C.C, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JBP
 
 | | Novel Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules | | Descriptor: | 1-(4-{2-[(6-{4-[2-(4-hydroxypiperidin-1-yl)ethoxy]phenyl}furo[2,3-d]pyrimidin-4-yl)amino]ethyl}phenyl)-3-phenylurea, Aurora Kinase A | | Authors: | Wu, J.S, Leou, J.S, Peng, Y.H, Hsueh, C.C, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LCS
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with hydantoin | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, ZINC ION, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S. | | Deposit date: | 2013-06-23 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
4LCR
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBA | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
5XTZ
 
 | | Crystal structure of GAS41 YEATS bound to H3K27ac peptide | | Descriptor: | ACETATE ION, THR-LYS-ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, YEATS domain-containing protein 4 | | Authors: | Li, H.T, Zhao, D. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity.
Cell Discov, 4, 2018
|
|
7EHJ
 
 | | human MTHFD2 in complex with compound 21, cofactor and phosphate. | | Descriptor: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Lee, L.C, Peng, Y.H, Wu, S.Y. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
7EHM
 
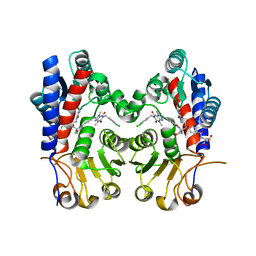 | | Human MTHFD2 in complex with compound 21 and 15 | | Descriptor: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, (2S)-2-[[4-[[1-[(3,4-dichlorophenyl)methyl]-3,7-dimethyl-2,6-bis(oxidanylidene)purin-8-yl]amino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Lee, L.C, Peng, Y.H, Wu, S.Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
7EHN
 
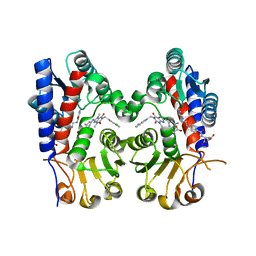 | | Human MTHFD2 in complex with compound 21 and 9 | | Descriptor: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, 3-[4-[[1-[(4-chloranyl-1H-indol-2-yl)methyl]-3,7-dimethyl-2,6-bis(oxidanylidene)purin-8-yl]amino]-6-methyl-pyrimidin-2-yl]propanoic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Lee, L.C, Peng, Y.H, Wu, S.Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
7EHV
 
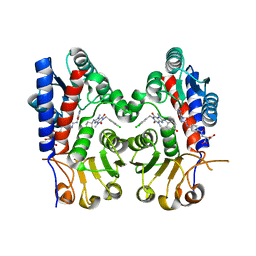 | | Human MTHFD2 in complex with compound 21 and 3 | | Descriptor: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, 1-(3,4-dichlorobenzyl)-8-(((1R,4R)-4-hydroxycyclohexyl)amino)-3,7-dimethyl-3,7-dihydro-1H-purine-2,6-dione, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Lee, L.C, Peng, Y.H, Wu, S.Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
6KOF
 
 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) in complex with compound 47 | | Descriptor: | 1-(4-cyanophenyl)-3-[[3-(2-cyclopropylethynyl)imidazo[2,1-b][1,3]thiazol-5-yl]methyl]thiourea, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, S.Y. | | Deposit date: | 2019-08-09 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Unique Sulfur-Aromatic Interactions Contribute to the Binding of Potent Imidazothiazole Indoleamine 2,3-Dioxygenase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6KW7
 
 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) in complex with compound 12 | | Descriptor: | 3-(4-bromophenyl)imidazo[2,1-b][1,3]thiazole, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, S.Y. | | Deposit date: | 2019-09-06 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Unique Sulfur-Aromatic Interactions Contribute to the Binding of Potent Imidazothiazole Indoleamine 2,3-Dioxygenase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6KPS
 
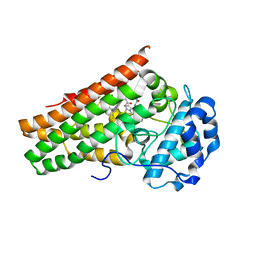 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) in complex with compound 36 | | Descriptor: | 1-(4-cyanophenyl)-3-[[3-(2-cyclopropylethynyl)imidazo[2,1-b][1,3]thiazol-5-yl]methyl]urea, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, S.Y. | | Deposit date: | 2019-08-16 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Unique Sulfur-Aromatic Interactions Contribute to the Binding of Potent Imidazothiazole Indoleamine 2,3-Dioxygenase Inhibitors.
J.Med.Chem., 63, 2020
|
|
5EK4
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-[(5~{S})-6-fluoranyl-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, S.Y, Peng, Y.H, Wu, J.S. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
5ETW
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-pyrido[3,4-b]indol-9-yl-ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, S.Y, Peng, Y.H, Wu, J.S. | | Deposit date: | 2015-11-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1.
J.Med.Chem., 59, 2016
|
|
5EK3
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-[(5~{S})-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
5EK2
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | 1-cyclohexyl-2-[(5~{S})-6-fluoranyl-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanone, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
