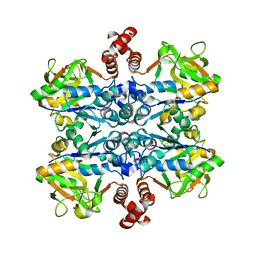
2E3D
 
 | |
2UDP
 
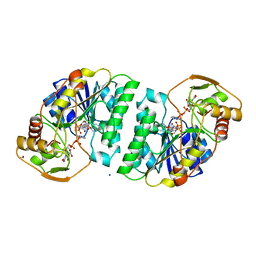 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHENYL-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray structure of UDP-galactose 4-epimerase complexed with UDP-phenol.
Protein Sci., 5, 1996
|
|
5KGH
 
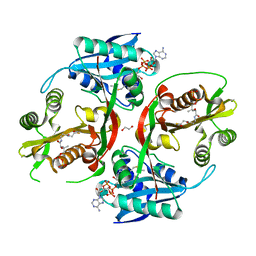 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant Y297F | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KF9
 
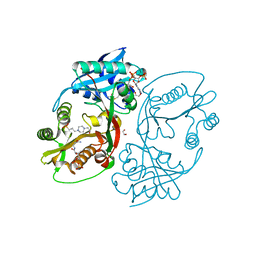 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Holden, H.M, Tipton, P.A. | | Deposit date: | 2016-06-12 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KGJ
 
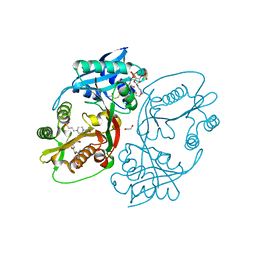 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with galactosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-alpha-D-galactopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KGA
 
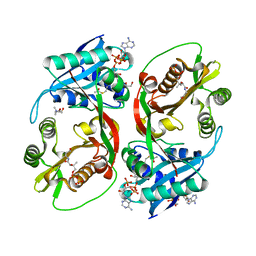 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant D287N, in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
4XD1
 
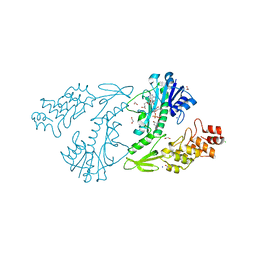 | | X-ray structure of the N-formyltransferase QdtF from Providencia alcalifaciens, W305A mutant, in the presence of TDP-Qui3N and N5-THF | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New Role for the Ankyrin Repeat Revealed by a Study of the N-Formyltransferase from Providencia alcalifaciens.
Biochemistry, 54, 2015
|
|
5KTC
 
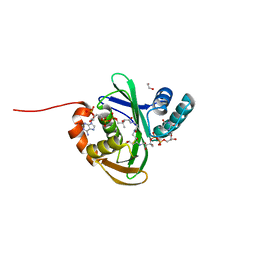 | | FdhC with bound products: Coenzyme A and 3-[(R)-3-hydroxybutanoylamino]-3,6-dideoxy-d-galactose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, FdhC, ... | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
4XD0
 
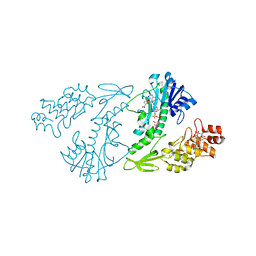 | | X-ray structure of the N-formyltransferase QdtF from Providencia alcalifaciens | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, CHLORIDE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New Role for the Ankyrin Repeat Revealed by a Study of the N-Formyltransferase from Providencia alcalifaciens.
Biochemistry, 54, 2015
|
|
4XCZ
 
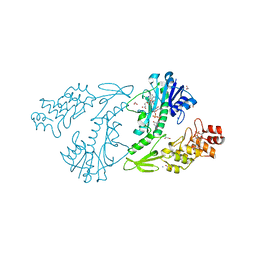 | | X-ray structure of the N-formyltransferase QdtF from Providencia alcalifaciens in complex with TDP-Qui3n and N5-THF | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New Role for the Ankyrin Repeat Revealed by a Study of the N-Formyltransferase from Providencia alcalifaciens.
Biochemistry, 54, 2015
|
|
5KTA
 
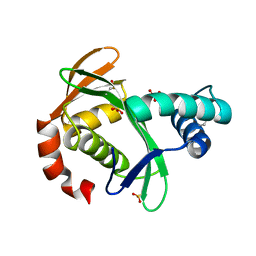 | | Apo FdhC- a nucleotide-linked sugar GNAT | | Descriptor: | DIMETHYL SULFOXIDE, FdhC, SULFATE ION | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
5KTD
 
 | | FdhC with bound products: Coenzyme A and dTDP-3-amino-3,6-dideoxy-d-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, FdhC, ... | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
4YFV
 
 | | X-ray structure of the 4-N-formyltransferase VioF from Providencia alcalifaciens O30 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, VioF | | Authors: | Genthe, N.A, Thoden, J.B, Benning, M.M, Holden, H.M. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular structure of an N-formyltransferase from Providencia alcalifaciens O30.
Protein Sci., 24, 2015
|
|
4YFY
 
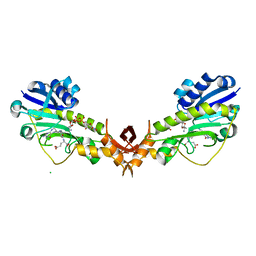 | | X-ray structure of the Viof N-formyltransferase from Providencia alcalifaciens O30 in complex with THF and TDP-Qui4N | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Genthe, N.A, Thoden, J.B, Benning, M.M, Holden, H.M. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structure of an N-formyltransferase from Providencia alcalifaciens O30.
Protein Sci., 24, 2015
|
|
4ZTC
 
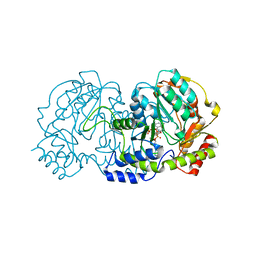 | | PglE Aminotransferase in complex with External Aldimine, Mutant K184A | | Descriptor: | Aminotransferase homolog, [(2R,3R,4R,5S,6R)-3-acetamido-6-methyl-5-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-4-oxidanyl-oxan-2-yl] [[(2R,3S,4R,5R)-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] hydrogen phosphate | | Authors: | Riegert, A.S, Thoden, J.B, Young, N.M, Watson, D.C, Holden, H.M. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the external aldimine form of PglE, an aminotransferase required for N,N'-diacetylbacillosamine biosynthesis.
Protein Sci., 24, 2015
|
|
4ZU5
 
 | | Crystal structure of the QdtA 3,4-Ketoisomerase from Thermoanaerobacterium thermosaccharolyticum, apo form | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, QdtA, THYMIDINE | | Authors: | Thoden, J.B, Vinogradov, E, Gilbert, M, Salinger, A.J, Holden, H.M. | | Deposit date: | 2015-05-15 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacterial Sugar 3,4-Ketoisomerases: Structural Insight into Product Stereochemistry.
Biochemistry, 54, 2015
|
|
4ZU7
 
 | | X-ray structure if the QdtA 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum, double mutant Y17R/R97H, in complex with TDP | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, QdtA, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Thoden, J.B, Vinogradov, E, Gilbert, M, Salinger, A.J, Holden, H.M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial Sugar 3,4-Ketoisomerases: Structural Insight into Product Stereochemistry.
Biochemistry, 54, 2015
|
|
4ZU4
 
 | | X-ray structure of the 3,4-ketoisomerase domain of FdtD from Shewanella denitrificans | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Thoden, J.B, Vinogradov, E, Gilbert, M, Salinger, A.J, Holden, H.M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacterial Sugar 3,4-Ketoisomerases: Structural Insight into Product Stereochemistry.
Biochemistry, 54, 2015
|
|
8SKP
 
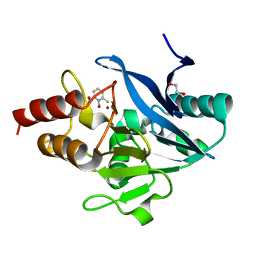 | | X-ray structure of the NDM-4 beta-lactamase from Klebsiella pneumonia in complex with 1-hydroxypyridine-2(1H)-thione-6-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 1-hydroxy-6-sulfanylidene-1,6-dihydropyridine-2-carboxylic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of a novel inhibitor for the New Delhi metallo-beta-lactamase-4: Implications for drug design and combating bacterial drug resistance.
J.Biol.Chem., 299, 2023
|
|
8SKO
 
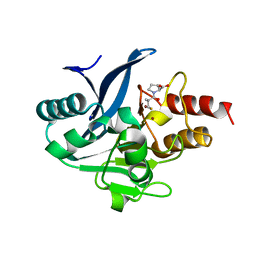 | |
8V4G
 
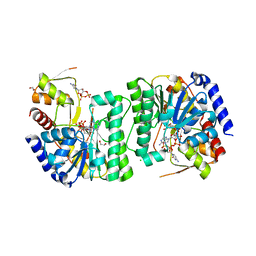 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP and NADP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Schumann, M.E, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
8V4H
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP-glucitol | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Putative nucleotide sugar dehydratase, ... | | Authors: | Thoden, J.B, Schumann, M.E, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
8SK2
 
 | |
8SY9
 
 | | X-ray crystal structure of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N variant in the presence of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid, UDP-N-acetylglucosamine and UDP at pH 7 | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SXY
 
 | | X-ray crystal structure of UDP- 2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27 in complex with its product UDP-2,3-diacetamido-2,3-dideoxy-d-mannuronic acid at pH 5 | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase | | Authors: | McKnight, J.O, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
