3FRK
 
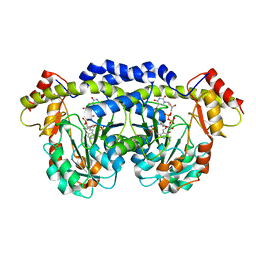 | | X-ray structure of QdtB from T. thermosaccharolyticum in complex with a PLP:TDP-3-aminoquinovose aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, QdtB | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of QdtB, an aminotransferase required for the biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose.
Biochemistry, 48, 2009
|
|
7S41
 
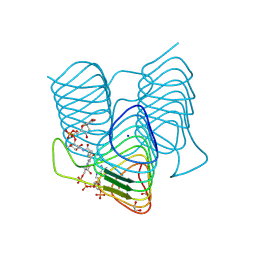 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-acetamido-3,6-dideoxy-D-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S3U
 
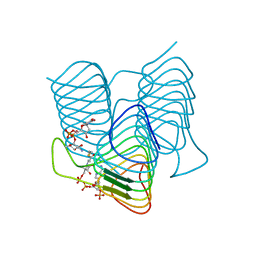 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S42
 
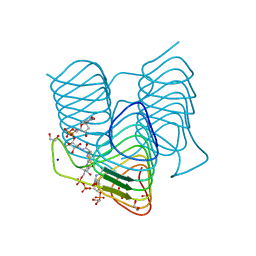 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-acetamido-3,6-dideoxy-D-galactose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S3W
 
 | | Crystal structure of an N-acetyltransferase from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S44
 
 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-galactose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S43
 
 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Coenzyme A and dTDP-3-amino-3,6-dideoxy-D-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-acetyltransferase, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
7S45
 
 | | Crystal structure of an N-acetyltransferase, C80T mutant, from Helicobacter pullorum in the presence of Acetyl Coenzyme A and dTDP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETYL COENZYME *A, ... | | Authors: | Griffiths, W.A, Spencer, K.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemical investigation of an N-acetyltransferase from Helicobacter pullorum.
Protein Sci., 30, 2021
|
|
3K5H
 
 | | Crystal structure of carboxyaminoimidazole ribonucleotide synthase from asperigillus clavatus complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphoribosyl-aminoimidazole carboxylase | | Authors: | Thoden, J.B, Holden, H.M, Paritala, H, Firestine, S.M. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of Aspergillus clavatus N(5)-carboxyaminoimidazole ribonucleotide synthetase
Biochemistry, 49, 2010
|
|
3K5I
 
 | | Crystal structure of N5-carboxyaminoimidazole synthase from aspergillus clavatus in complex with ADP and 5-aminoimadazole ribonucleotide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Holden, H.M, Paritala, H, Firestine, S.M. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional studies of Aspergillus clavatus N(5)-carboxyaminoimidazole ribonucleotide synthetase
Biochemistry, 49, 2010
|
|
2MYS
 
 | |
6V33
 
 | | X-ray structure of a sugar N-formyltransferase from Pseudomonas congelans | | Descriptor: | 1,2-ETHANEDIOL, FOLIC ACID, dTDP-4-amino-4,6-dideoxyglucose, ... | | Authors: | Girardi, N.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Misannotations of the genes encoding sugar N-formyltransferases.
Protein Sci., 29, 2020
|
|
6V2T
 
 | | X-ray structure of a sugar N-formyltransferase from Shewanella sp FDAARGOS_354 | | Descriptor: | 1,2-ETHANEDIOL, FOLIC ACID, PHOSPHATE ION, ... | | Authors: | Girardi, N.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Misannotations of the genes encoding sugar N-formyltransferases.
Protein Sci., 29, 2020
|
|
6VLO
 
 | | X-ray Structure of the R141 Sugar 4,6-dehydratase from Acanthamoeba polyphaga Minivirus | | Descriptor: | NICKEL (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative dTDP-D-glucose 4,6-dehydratase, ... | | Authors: | Thoden, J.B, Ferek, J.D, Holden, H.M. | | Deposit date: | 2020-01-24 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical analysis of a sugar 4,6-dehydratase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 29, 2020
|
|
6VO6
 
 | | Crystal Structure of Cj1427, an Essential NAD-dependent Dehydrogenase from Campylobacter jejuni, in the Presence of NADH and GDP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Anderson, T.K, Spencer, K.D, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
6VO8
 
 | | X-ray structure of the Cj1427 in the presence of NADH and GDP-D-glycero-D-mannoheptose, an essential NAD-dependent dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative sugar-nucleotide epimerase/dehydratease, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S},5~{S},6~{S})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Spencer, K.D, Anderson, T.K, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
3B8X
 
 | | Crystal structure of GDP-4-keto-6-deoxymannose-3-dehydratase (ColD) H188N mutant with bound GDP-perosamine | | Descriptor: | 1,2-ETHANEDIOL, Pyridoxamine 5-phosphate-dependent dehydrase, SODIUM ION, ... | | Authors: | Cook, P.D, Holden, H.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GDP-4-Keto-6-deoxy-D-mannose 3-Dehydratase, Accommodating a Sugar Substrate in the Active Site.
J.Biol.Chem., 283, 2008
|
|
3BN1
 
 | | Crystal structure of GDP-perosamine synthase | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, Perosamine synthetase, ... | | Authors: | Cook, P.D, Holden, H.M. | | Deposit date: | 2007-12-13 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GDP-Perosamine Synthase: Structural Analysis and Production of a Novel Trideoxysugar
Biochemistry, 47, 2008
|
|
3E1K
 
 | |
7MFO
 
 | | X-ray structure of the L136 Aminotransferase from Acanthamoeba polyphaga mimivirus in the presence of TDP and PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Ferek, J.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7MFP
 
 | | crystal structure of the L136 aminotransferase K185A from acanthamoeba polyphaga mimivirus in the presence of the UDP-viosamine external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-dihydroxy-5-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-methyloxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7MFQ
 
 | | crystal structure of the L136 aminotransferase from acanthamoeba polyphaga mimivirus in complex with the TDP-viosamine external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-DIHYDROXY-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)IMINO]-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL [(2R,3S,5R)-3-HYDROXY-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7N63
 
 | | X-ray structure of HCAN_0200, an aminotransferase from Helicobacter canadensis in complex with its external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Griffiths, W.A, Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N7C
 
 | | crystal structure of the N-formyltransferase HCAN_0200 from helicobacter canadensis in complex with folinic acid and dTDP-3-aminoquinovose | | Descriptor: | 1,2-ETHANEDIOL, Formyl_trans_N domain-containing protein, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
7N7B
 
 | | crystal structure of the N-formyltrasferase HCAN_0200 from Helicobacter canadensis on complex with folinic acid and dTDP-3-aminofucose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Heisdorf, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
