2MOV
 
 | | Receptor for Advanced Glycation End Products (RAGE) Specifically Recognizes Methylglyoxal Derived AGEs. | | Descriptor: | Advanced glycosylation end product-specific receptor, N~5~-[(5R)-5-methyl-4-oxo-4,5-dihydro-1H-imidazol-2-yl]-L-ornithine | | Authors: | Shekhtman, A, Xue, J, Ray, R, Singer, D, Bohme, D, Burz, D.S, Rai, V, Hoffman, R. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Receptor for Advanced Glycation End Products (RAGE) Specifically Recognizes Methylglyoxal-Derived AGEs.
Biochemistry, 53, 2014
|
|
2L7U
 
 | | Structure of CEL-PEP-RAGE V domain complex | | Descriptor: | Advanced glycosylation end product-specific receptor, Serum albumin peptide | | Authors: | Xue, J, Rai, V, Schmidt, A, Frolov, S, Reverdatto, S, Singer, D, Chabierski, S, Xie, J, Burz, D, Shekhtman, A, Hoffman, R. | | Deposit date: | 2010-12-23 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Advanced glycation end product recognition by the receptor for AGEs.
Structure, 19, 2011
|
|
2V4L
 
 | | complex of human phosphoinositide 3-kinase catalytic subunit gamma (p110 gamma) with PIK-284 | | Descriptor: | 3-[4-AMINO-1-(1-METHYLETHYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL]PHENOL, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Apsel, B, Gonzalez, B, Blair, J.A, Nazif, T.M, Feldman, M.E, Williams, R.L, Shokat, K.M, Knight, Z.A. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeted Polypharmacology: Discovery of Dual Inhibitors of Tyrosine and Phosphoinositide Kinases.
Nat.Chem.Biol., 4, 2008
|
|
3ENE
 
 | | Complex of PI3K gamma with an inhibitor | | Descriptor: | 1-methyl-3-naphthalen-2-yl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Apsel, B, Blair, J.A, Gonzalez, B.Z, Nazif, T.M, Feldman, M.E, Williams, R.L, Shokat, K.M, Knight, Z.A. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases
Nat.Chem.Biol., 4, 2008
|
|
3EN5
 
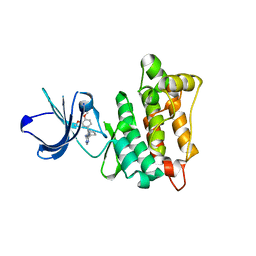 | | Targeted polypharmacology: crystal structure of the c-Src kinase domain in complex with PP494, a multitargeted kinase inhibitor | | Descriptor: | 1-cyclobutyl-3-(3,4-dimethoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Blair, J.A, Apsel, B, Knight, Z.A, Shokat, K.M. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases.
Nat.Chem.Biol., 4, 2008
|
|
3EN6
 
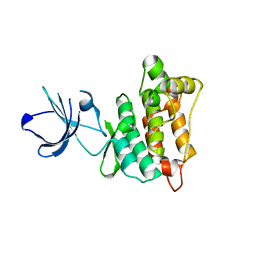 | | Targeted polypharmacology: crystal structure of the c-Src kinase domain in complex with PP102, a multitargeted kinase inhibitor | | Descriptor: | 1-(1-methylethyl)-3-quinolin-6-yl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Blair, J.A, Apsel, B, Knight, Z.A, Shokat, K.M. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases.
Nat.Chem.Biol., 4, 2008
|
|
3EN7
 
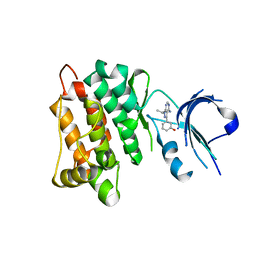 | | Targeted polypharmacology: crystal structure of the c-Src kinase domain in complex with S1, a multitargeted kinase inhibitor | | Descriptor: | 3-[4-AMINO-1-(1-METHYLETHYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL]PHENOL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Blair, J.A, Apsel, B, Knight, Z.A, Shokat, K.M. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases.
Nat.Chem.Biol., 4, 2008
|
|
3EN4
 
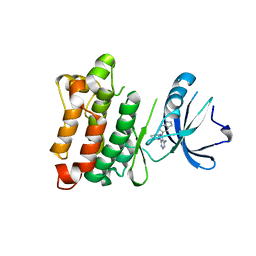 | | Targeted polypharmacology: crystal structure of the c-Src kinase domain in complex with PP121, a multitargeted kinase inhibitor | | Descriptor: | 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Blair, J.A, Apsel, B, Knight, Z.A, Shokat, K.M. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases.
Nat.Chem.Biol., 4, 2008
|
|
4O36
 
 | | Semisynthetic RNase S1-15-H7/11-Q10 | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, S-peptide, ... | | Authors: | Genz, M, Strater, N. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | An Artificial Imine Reductase based on the Ribonuclease S scaffold
Chem.Cat.Chem, 2014
|
|
4EZP
 
 | |
4F01
 
 | |
4EZT
 
 | |
4EZR
 
 | |
4EZW
 
 | |
4EZO
 
 | |
4EZZ
 
 | |
4F00
 
 | |
4EZQ
 
 | |
3O1W
 
 | | Crystal structure of dimeric KlHxk1 in crystal form III | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O1B
 
 | | CRYSTAL STRUCTURE OF DIMERIC KLHXK1 IN CRYSTAL FORM II | | Descriptor: | Hexokinase | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-21 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O5B
 
 | | Crystal structure of dimeric KlHxk1 in crystal form VII with glucose bound (open state) | | Descriptor: | Hexokinase, SULFATE ION, beta-D-glucopyranose | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O6W
 
 | | Crystal structure of monomeric KlHxk1 in crystal form VIII (open state) | | Descriptor: | GLYCEROL, Hexokinase, PHOSPHATE ION | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O4W
 
 | | Crystal structure of dimeric KlHxk1 in crystal form IV | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-27 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O08
 
 | | Crystal structure of dimeric KlHxk1 in crystal form I | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hexokinase, SULFATE ION | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-19 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O8M
 
 | | Crystal structure of monomeric KlHxk1 in crystal form XI with glucose bound (closed state) | | Descriptor: | CHLORIDE ION, Hexokinase, alpha-D-glucopyranose, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
