3TED
 
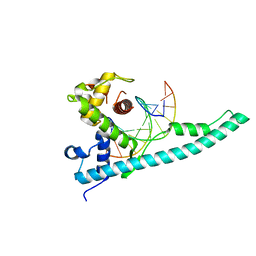 | | Crystal structure of the Chd1 DNA-binding domain in complex with a DNA duplex | | Descriptor: | 5'-D(*CP*CP*AP*TP*AP*TP*AP*TP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*AP*TP*AP*TP*AP*TP*GP*G)-3', Chromo domain-containing protein 1 | | Authors: | Sharma, A, Jenkins, K.R, Heroux, A, Bowman, G.D. | | Deposit date: | 2011-08-12 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA-binding domain of Chd1 in complex with a DNA duplex
J.Biol.Chem., 2011
|
|
4POC
 
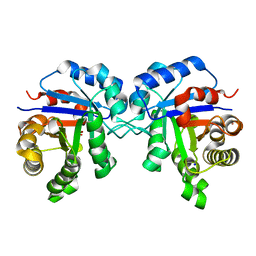 | | Structure of Triosephosphate Isomerase Wild Type human enzyme. | | Descriptor: | BROMIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Amrich, C.G, Aslam, A.A, Heroux, A, VanDemark, A.P. | | Deposit date: | 2014-02-25 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency.
Biochim.Biophys.Acta, 1852, 2015
|
|
4POD
 
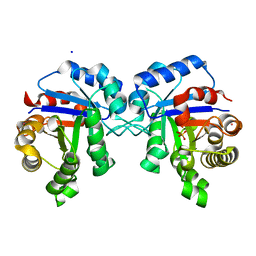 | | Structure of Triosephosphate Isomerase I170V mutant human enzyme. | | Descriptor: | BROMIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Amrich, C.G, Aslam, A.A, Heroux, A, VanDemark, A.P. | | Deposit date: | 2014-02-25 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency.
Biochim.Biophys.Acta, 1852, 2015
|
|
3BIP
 
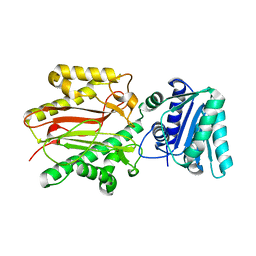 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | FACT complex subunit SPT16 | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
3BIT
 
 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, FACT complex subunit SPT16, ... | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
3BIQ
 
 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | FACT complex subunit SPT16, GLYCEROL | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
3BVH
 
 | | Crystal Structure of Recombinant gammaD364A Fibrinogen Fragment D with the Peptide Ligand Gly-Pro-Arg-Pro-Amide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-mer peptide GPRP, CALCIUM ION, ... | | Authors: | Bowley, S.R, Merenbloom, B.K, Betts, L, Okumura, N, Heroux, A, Gorkun, O.V, Lord, S.T. | | Deposit date: | 2008-01-07 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Polymerization-defective fibrinogen variant gammaD364A binds knob "A" peptide mimic.
Biochemistry, 47, 2008
|
|
3CDG
 
 | | Human CD94/NKG2A in complex with HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Petrie, E.J, Clements, C.S, Lin, J, Sullivan, L.C, Johnson, D, Huyton, T, Heroux, A, Hoare, H.L, Beddoe, T, Reid, H.H, Wilce, M.C.J, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence
J.Exp.Med., 205, 2008
|
|
3D0X
 
 | |
3D0U
 
 | | Crystal Structure of Lysine Riboswitch Bound to Lysine | | Descriptor: | IRIDIUM HEXAMMINE ION, LYSINE, Lysine Riboswitch RNA | | Authors: | Garst, A.D, Heroux, A, Rambo, R.P, Batey, R.T. | | Deposit date: | 2008-05-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the lysine riboswitch regulatory mRNA element.
J.Biol.Chem., 283, 2008
|
|
3D7J
 
 | | SCO6650, a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein SCO6650 | | Authors: | Spoonamore, J.E, Roberts, S.A, Heroux, A, Bandarian, V. | | Deposit date: | 2008-05-21 | | Release date: | 2008-10-21 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3E66
 
 | | Crystal structure of the beta-finger domain of yeast Prp8 | | Descriptor: | PRP8 | | Authors: | Yang, K, Zhang, L, Xu, T, Heroux, A, Zhao, R. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3EIH
 
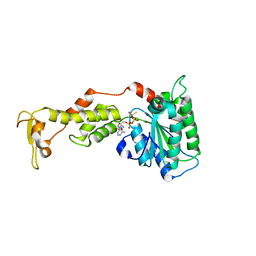 | | Crystal structure of S.cerevisiae Vps4 in the presence of ATPgammaS | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gonciarz, M.D, Whitby, F.G, Eckert, D.M, Kieffer, C, Heroux, A, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Biochemical and structural studies of yeast vps4 oligomerization.
J.Mol.Biol., 384, 2008
|
|
3EIE
 
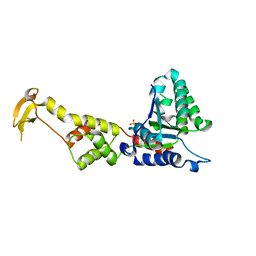 | | Crystal Structure of S.cerevisiae Vps4 in the SO4-bound state | | Descriptor: | SULFATE ION, Vacuolar protein sorting-associated protein 4 | | Authors: | Gonciarz, M.D, Whitby, F.G, Eckert, D.M, Kieffer, C, Heroux, A, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural studies of yeast vps4 oligomerization.
J.Mol.Biol., 384, 2008
|
|
3O48
 
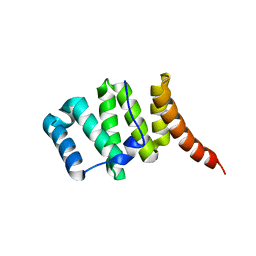 | | Crystal structure of fission protein Fis1 from Saccharomyces cerevisiae | | Descriptor: | Mitochondria fission 1 protein | | Authors: | Tooley, J.E, Khangulov, V, Heroux, A, Bosch, J, Hill, R.B. | | Deposit date: | 2010-07-26 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.75 Angstrom resolution structure of fission protein Fis1 from Saccharomyces cerevisiae reveals elusive interactions of the autoinhibitory domain
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3PA6
 
 | |
3OWF
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66R at cryogenic temperature | | Descriptor: | CALCIUM ION, PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, ... | | Authors: | Schlessman, J.L, Khangulov, V, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2010-09-17 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Domain swapping promoted by a single mutation that introduces an ionizable group into the hydrophobic core of a protein
To be Published
|
|
3QB3
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92KL25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sue, G, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2011-01-12 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Relocation of internal ionizable residues to cavities created by single alanine substitutions
To be Published
|
|
3QOJ
 
 | | Cryogenic structure of Staphylococcal nuclease variant D+PHS/V23K | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2011-02-10 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determinants of pKa values of internal ionizable groups: properties of ion pairs in the protein core
To be Published
|
|
3QZ0
 
 | | Structure of Treponema denticola Factor H Binding protein (FhbB), selenomethionine derivative | | Descriptor: | Factor H binding protein, GLYCEROL, THIOCYANATE ION | | Authors: | Miller, D.P, McDowell, J.V, Heroux, A, Bell, J.K, Marconi, R.T, Conrad, D.H, Burgner, J.W. | | Deposit date: | 2011-03-04 | | Release date: | 2012-03-07 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of factor H-binding protein B (FhbB) of the periopathogen, Treponema denticola: insights into progression of periodontal disease.
J.Biol.Chem., 287, 2012
|
|
3R15
 
 | | Structure Treponema Denticola Factor H Binding Protein | | Descriptor: | Factor H binding protein, THIOCYANATE ION | | Authors: | Miller, D.P, McDowell, J.V, Burgner, J, Heroux, A, Bell, J.K, Marconi, R.T. | | Deposit date: | 2011-03-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure Treponema Denticola Factor H Binding Protein
To be Published
|
|
3RYV
 
 | | Carbonic Anhydrase complexed with N-ethyl-4-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, N-ethyl-4-sulfamoylbenzamide, ZINC ION | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RZ1
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-(2,2,3,3,4,4,5,5,5-nonafluoropentyl)-4-sulfamoylbenzamide, ... | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RYJ
 
 | | Carbonic Anhydrase complexed with 4-sulfamoyl-N-(2,2,2-trifluoroethyl)benzamide | | Descriptor: | 4-sulfamoyl-N-(2,2,2-trifluoroethyl)benzamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RZ5
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-pentyl-4-sulfamoylbenzamide, ... | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
