5EOS
 
 | |
5W6W
 
 | |
9GS2
 
 | |
9GSN
 
 | |
9GU9
 
 | |
8UY5
 
 | | [ZP] Self-assembling DNA crystal with expanded genetic code using C,A,T,G, Z and P nucleotides | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*(DP)P*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*(DZ)P*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Sha, R, Ohayon, Y.P, Hernandez, C. | | Deposit date: | 2023-11-13 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.562 Å) | | Cite: | Six-Letter DNA Nanotechnology: Incorporation of Z - P Base Pairs into Self-Assembling 3D Crystals.
Nano Lett., 24, 2024
|
|
6EU6
 
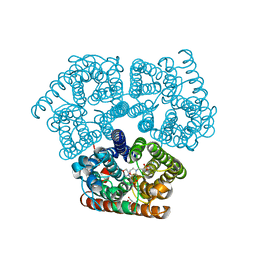 | | Sensor Amt Protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMMONIUM ION, Ammonium transporter, ... | | Authors: | Pflueger, T, Hernandez, C, Andrade, S.L.A. | | Deposit date: | 2017-10-28 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun, 9, 2018
|
|
8WKZ
 
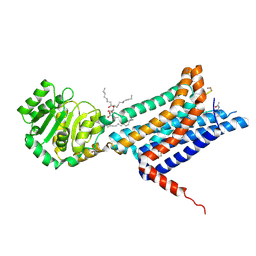 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S31 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
8WKY
 
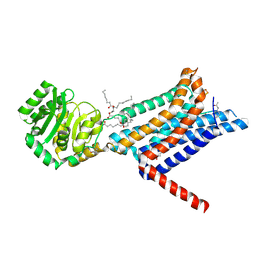 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S25 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, N-(2-aminoethyl)-5-(2-{[4-(morpholin-4-yl)pyridin-2-yl]amino}-1,3-thiazol-5-yl)pyridine-3-carboxamide, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9NAA
 
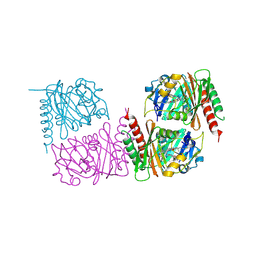 | | Unusual structure of a bacteriophytochrome fragment derived from full length SaBphP2 | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, histidine kinase | | Authors: | Schmidt, M, Prabin, K. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of myxobacterial phytochrome revealed by cryo-EM using the Spotiton technique and with x-ray crystallography.
Struct Dyn., 12, 2025
|
|
9H80
 
 | | Structure of the outer membrane exopolysaccharide transporter PelBC | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, PelB, PelC | | Authors: | Benedens, M, Rosales, C, Beckmann, R, Kedrov, A. | | Deposit date: | 2024-10-28 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Assembly and the gating mechanism of the Pel exopolysaccharide export complex PelBC of Pseudomonas aeruginosa.
Nat Commun, 16, 2025
|
|
7P0M
 
 | |
7P09
 
 | |
7P0B
 
 | |
5I36
 
 | | Crystal structure of color device state A | | Descriptor: | DNA (26-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*GP*TP*GP*GP*TP*AP*TP*C)-3'), DNA (5'-D(*CP*AP*GP*AP*TP*AP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Hao, Y, Kristiansen, M, Sha, R, Birktoft, J, Mao, C, Seeman, N.C. | | Deposit date: | 2016-02-09 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.123 Å) | | Cite: | A device that operates within a self-assembled 3D DNA crystal.
Nat Chem, 9, 2017
|
|
5KEK
 
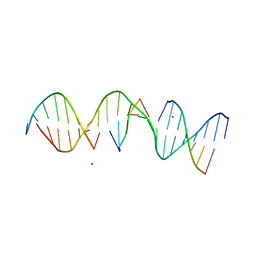 | | Structure Determination of a Self-Assembling DNA Crystal | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, Birktoft, J.J, Seeman, N.C, Yan, H. | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Construction and Structure Determination of a Three-Dimensional DNA Crystal.
J.Am.Chem.Soc., 138, 2016
|
|
5KEO
 
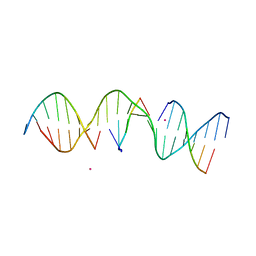 | | Structure Determination of a Self-Assembling DNA Crystal | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, Birktoft, J.J, Seeman, N.C, Yan, H. | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Construction and Structure Determination of a Three-Dimensional DNA Crystal.
J.Am.Chem.Soc., 138, 2016
|
|
5I6Q
 
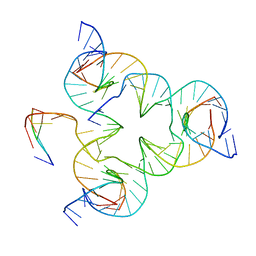 | | Crystal structure of color device state B | | Descriptor: | DNA (26-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*GP*TP*GP*GP*TP*AP*TP*C)-3'), DNA (5'-D(*CP*AP*GP*AP*TP*AP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Hao, Y, Birktoft, J, Seeman, N.C. | | Deposit date: | 2016-02-16 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.912 Å) | | Cite: | A device that operates within a self-assembled 3D DNA crystal.
Nat Chem, 9, 2017
|
|
5I6T
 
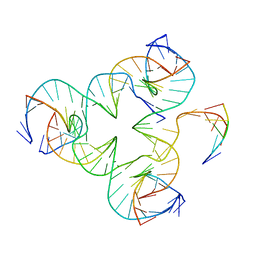 | | Crystal structure of color device state C | | Descriptor: | DNA (26-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*GP*TP*GP*GP*TP*AP*TP*C)-3'), DNA (5'-D(*CP*AP*GP*AP*TP*AP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Hao, Y, Birktoft, J, Seeman, N.C. | | Deposit date: | 2016-02-16 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.283 Å) | | Cite: | A device that operates within a self-assembled 3D DNA crystal.
Nat Chem, 9, 2017
|
|
9GGC
 
 | |
9GGB
 
 | |
9GGF
 
 | |
9GGE
 
 | |
9GGD
 
 | |
7KRN
 
 | | Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
