7R3H
 
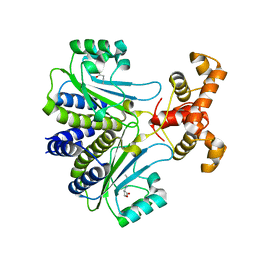 | |
7R3G
 
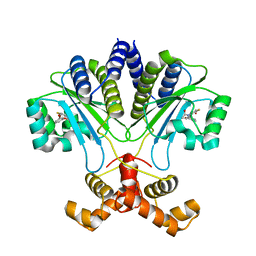 | |
7R3I
 
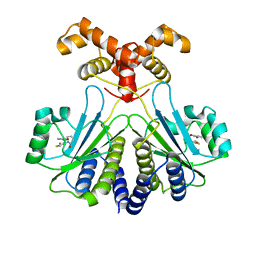 | |
7QY3
 
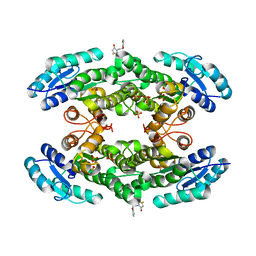 | | Crystal structure of the halohydrin dehalogenase HheG D114C mutant cross-linked with BMOE | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), Putative oxidoreductase, SULFATE ION | | Authors: | Henke, S, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Biocatalytically active and stable cross-linked enzyme crystals of halohydrin dehalogenase HheG by protein engineering
Chemcatchem, 2022
|
|
6TP9
 
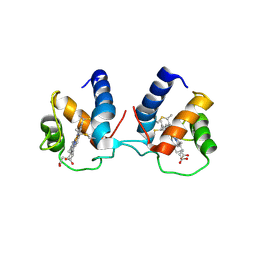 | | c-type cytochrome NirC | | Descriptor: | Cytochrome c55X, HEME C | | Authors: | Kluenemann, T, Henke, S, Blankenfeldt, W. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of the heme d1biosynthesis-associated small c-type cytochrome NirC reveals mixed oligomeric states in crystallo.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5VSZ
 
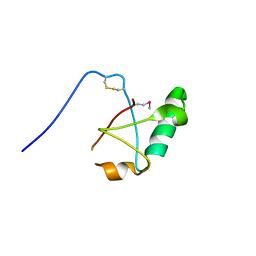 | |
5VSX
 
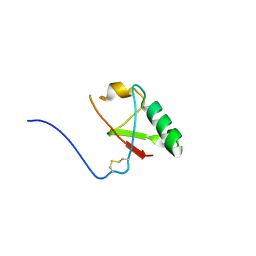 | |
8B4A
 
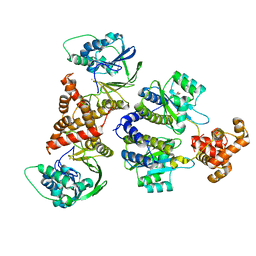 | | Nativ complex of PqsE and RhlR with autoinducer C4-HSL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Borgert, S.R, Blankenfeldt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
3T3C
 
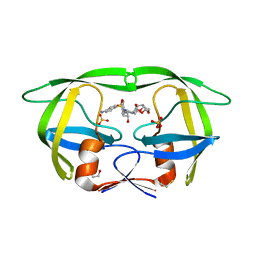 | | Structure of HIV PR resistant patient derived mutant (comprising 22 mutations) in complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, BETA-MERCAPTOETHANOL, HIV-1 protease, ... | | Authors: | Rezacova, P, Kozisek, M, Konvalinka, J, Saskova, K.G. | | Deposit date: | 2011-07-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations in HIV-1 gag and pol Compensate for the Loss of Viral Fitness Caused by a Highly Mutated Protease.
Antimicrob.Agents Chemother., 56, 2012
|
|
7R3J
 
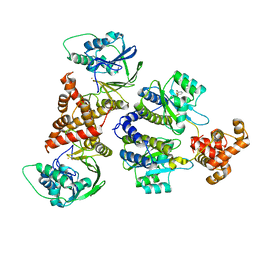 | | Nativ complex of PqsE and RhlR with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3F
 
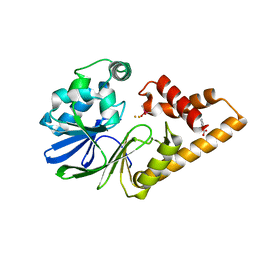 | | Monomeric PqsE mutant E187R | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, BENZOIC ACID, CACODYLATE ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3E
 
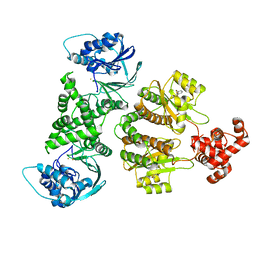 | | Fusion construct of PqsE and RhlR in complex with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase,Regulatory protein RhlR, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
1BDK
 
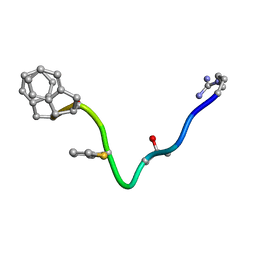 | | AN NMR, CD, MOLECULAR DYNAMICS, AND FLUOROMETRIC STUDY OF THE CONFORMATION OF THE BRADYKININ ANTAGONIST B-9340 IN WATER AND IN AQUEOUS MICELLAR SOLUTIONS | | Descriptor: | bradykinin antagonist B-9340 | | Authors: | Sejbal, J, Kotovych, G, Cann, J.R, Stewart, J.M, Gera, L. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-07 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | An NMR, CD, molecular dynamics, and fluorometric study of the conformation of the bradykinin antagonist B-9340 in water and in aqueous micellar solutions.
J.Med.Chem., 39, 1996
|
|
5KES
 
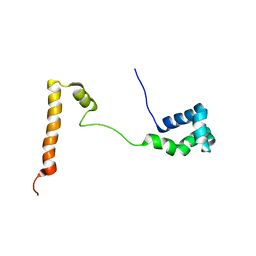 | | Solution structure of the yeast Ddi1 HDD domain | | Descriptor: | DNA damage-inducible protein 1 | | Authors: | Trempe, J.-F, Ratcliffe, C, Veverka, V, Saskova, K, Gehring, K. | | Deposit date: | 2016-06-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family.
Sci Rep, 6, 2016
|
|
5V45
 
 | |
5V46
 
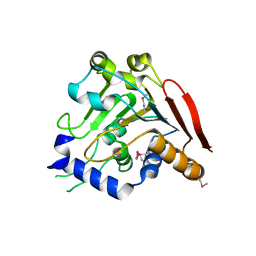 | | Crystal structure of the I113M, F270M, K291M, L308M mutant of SR1 domain of human sacsin | | Descriptor: | Sacsin | | Authors: | Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
5V44
 
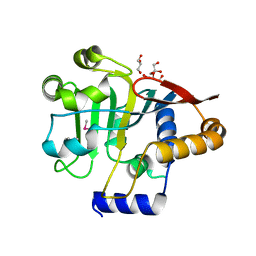 | |
5V47
 
 | | Crystal structure of the SR1 domain of lizard sacsin | | Descriptor: | Lizard sacsin, SULFATE ION | | Authors: | Pan, T, Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
4Z2Z
 
 | |
2N7E
 
 | |
