5TAW
 
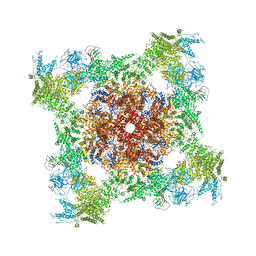 | | Structure of rabbit RyR1 (ryanodine dataset, all particles) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAL
 
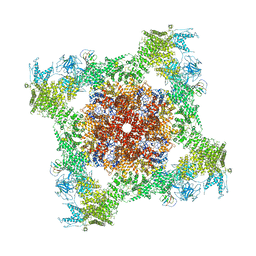 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 1&2) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAM
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 4) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T9M
 
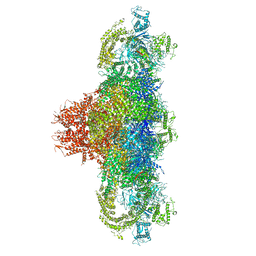 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 1) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAY
 
 | | Structure of rabbit RyR1 (ryanodine dataset, class 2) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T9S
 
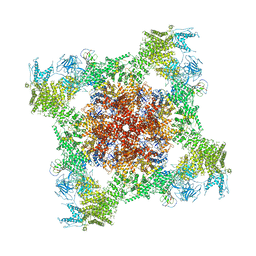 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 4) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB1
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, class 1) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T9R
 
 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 3) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB0
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, all particles) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAS
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TA3
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 2) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB2
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, class 2) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAN
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 3) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB3
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, class 3) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T9V
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAX
 
 | | Structure of rabbit RyR1 (ryanodine dataset, class 1) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5WUF
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | CADMIUM ION, Putative membrane protein | | Authors: | Mao, Y, Gao, F, Su, M, Wang, X.H, Zeng, Y, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUC
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SODIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUD
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUE
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
3LID
 
 | |
3LI9
 
 | |
3LI8
 
 | |
3LCK
 
 | |
3LIA
 
 | |
