7B8S
 
 | | Fusidic acid bound structure of bacterial efflux pump. | | Descriptor: | DARPin, FUSIDIC ACID, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Wilhelm, J, Sjuts, H, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
7B8T
 
 | | Levofloxacin bound structure of bacterial efflux pump. | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Wilhelm, J, Sjuts, H, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
7B8R
 
 | | Doxycycline bound structure of bacterial efflux pump. | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, DARPin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wilhelm, J, Sjuts, H, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
6RWV
 
 | | Structure of apo-LmCpfC | | Descriptor: | Ferrochelatase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6386379 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
6SV3
 
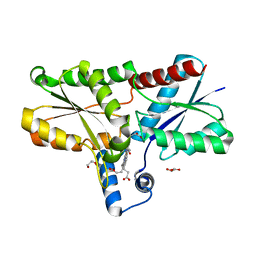 | | Structure of coproheme-LmCpfC | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Ferrochelatase, GLYCEROL | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-09-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64000869 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
4MSU
 
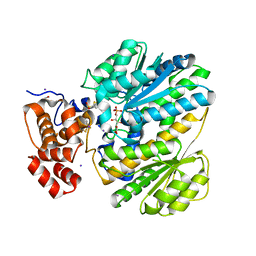 | | Human GKRP bound to AMG-6861 and Sorbitol-6-phosphate | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-{4-[4-(thiophen-2-ylsulfonyl)piperazin-1-yl]phenyl}propan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Ashton, K.S, Andrews, K.L, Bryan, M.C, Chen, J, Chen, K, Chen, M, Chmait, S, Croghan, M, Cupples, R, Fotsch, C, Helmering, J, Jordan, S.R, Kurzeja, R.J, Michelsen, K, Pennington, L.D, Poon, S.F, Sivits, G, Van, G, Vonderfecht, S.L, Wahl, R.C, Zhang, J, Lloyd, D.J, Hale, C, St Jean, D.J. | | Deposit date: | 2013-09-18 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Small Molecule Disruptors of the Glucokinase-Glucokinase Regulatory Protein Interaction: 1. Discovery of a Novel Tool Compound for in Vivo Proof-of-Concept.
J.Med.Chem., 57, 2014
|
|
4MQU
 
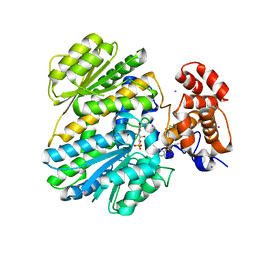 | | Human GKRP complexed to AMG-3969 and S6P | | Descriptor: | 2-{4-[(2S)-4-[(6-aminopyridin-3-yl)sulfonyl]-2-(prop-1-yn-1-yl)piperazin-1-yl]phenyl}-1,1,1,3,3,3-hexafluoropropan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | St Jean, D.J, Ashton, K.S, Bartberger, M.D, Chen, J, Chmait, S, Cupples, R, Galbreath, E, Helmering, J, Jordan, S.R, Liu, L. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 2. Leveraging structure-based drug design to identify analogues with improved pharmacokinetic profiles.
J.Med.Chem., 57, 2014
|
|
4MRO
 
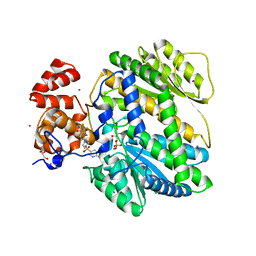 | | Human GKRP bound to AMG-5980 and S6P | | Descriptor: | 2-(4-{4-[(6-aminopyridin-3-yl)sulfonyl]piperazin-1-yl}phenyl)-1,1,1,3,3,3-hexafluoropropan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | St Jean, D.J, Ashton, K.S, Bartberger, M.D, Chen, J, Chmait, S, Cupples, R, Galbreath, E, Helmering, J, Jordan, S.R, Liu, L, Kunz, K, Michelsen, K, Nishimura, N, Pennington, L.D, Poon, S.F, Sivits, G, Stec, M.M, Tamayo, N, Van, G, Yang, K, Norman, M.H, Fotsch, C, LLoyd, D.J, Hale, C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 2. Leveraging structure-based drug design to identify analogues with improved pharmacokinetic profiles.
J.Med.Chem., 57, 2014
|
|
2K2G
 
 | | Solution structure of the wild-type catalytic domain of human matrix metalloproteinase 12 (MMP-12) in complex with a tight-binding inhibitor | | Descriptor: | Macrophage metalloelastase, N-(dibenzo[b,d]thiophen-3-ylsulfonyl)-L-valine, ZINC ION | | Authors: | Markus, M.A, Dwyer, B, Wolfrom, S, Li, J, Li, W, Malakian, K, Wilhelm, J, Tsao, D.H.H. | | Deposit date: | 2008-04-01 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of wild-type human matrix metalloproteinase 12 (MMP-12) in complex with a tight-binding inhibitor.
J.Biomol.Nmr, 41, 2008
|
|
3K1K
 
 | | Green fluorescent protein bound to enhancer nanobody | | Descriptor: | Enhancer, Green Fluorescent Protein | | Authors: | Kirchhofer, A, Helma, J, Schmidthals, K, Frauer, C, Cui, S, Karcher, A, Pellis, M, Muyldermans, S, Delucci, C.C, Cardoso, M.C, Leonhardt, H, Hopfner, K.-P, Rothbauer, U. | | Deposit date: | 2009-09-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Modulation of protein properties in living cells using nanobodies
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XXC
 
 | | Crystal structure of a camelid VHH raised against the HIV-1 capsid protein C-terminal domain. | | Descriptor: | CAMELID VHH 9 | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
2XT1
 
 | | Crystal structure of the HIV-1 capsid protein C-terminal domain (146- 231) in complex with a camelid VHH. | | Descriptor: | CAMELID VHH 9, GAG POLYPROTEIN, GLYCEROL | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-10-03 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
2XXM
 
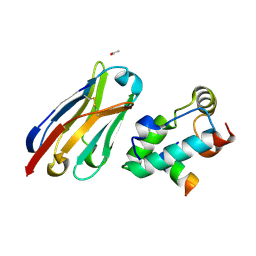 | | Crystal structure of the HIV-1 capsid protein C-terminal domain in complex with a camelid VHH and the CAI peptide. | | Descriptor: | ACETATE ION, CAMELID VHH 9, CAPSID PROTEIN P24, ... | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
2XV6
 
 | | Crystal structure of the HIV-1 capsid protein C-terminal domain (146- 220) in complex with a camelid VHH. | | Descriptor: | CAMELID VHH 9, CAPSID PROTEIN P24 | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
1IJZ
 
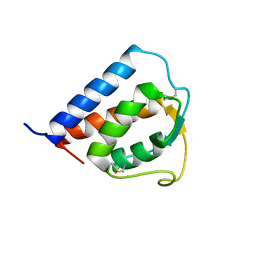 | |
1IK0
 
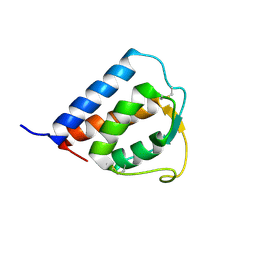 | |
3G9A
 
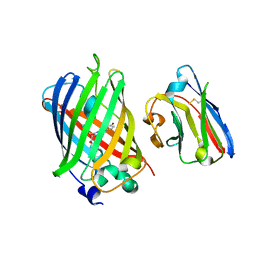 | | Green fluorescent protein bound to minimizer nanobody | | Descriptor: | Green fluorescent protein, Minimizer | | Authors: | Kirchhofer, A, Helma, J, Schmidthals, K, Frauer, C, Cui, S, Karcher, A, Pellis, M, Muyldermans, S, Delucci, C.C, Cardoso, M.C, Leonhardt, H, Hopfner, K.-P, Rothbauer, U. | | Deposit date: | 2009-02-13 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | Modulation of protein properties in living cells using nanobodies
Nat.Struct.Mol.Biol., 17, 2010
|
|
6Y7A
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with a chloroalkane-tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-02-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
6Y8P
 
 | | Crystal structure of SNAP-tag labeled with a benzyl-tetramethylrhodamine fluorophore | | Descriptor: | 1,2-ETHANEDIOL, O6-alkylguanine-DNA alkyltransferase mutant, ZINC ION, ... | | Authors: | Gotthard, G, Tanzer, T, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
6Y7B
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with a chloroalkane-carbopyronine fluorophore substrate | | Descriptor: | 4-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]-2-[3-(dimethylamino)-6-(dimethyl-$l^{4}-azanylidene)-10,10-dimethyl-anthracen-9-yl]benzoic acid, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-02-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
6ZCC
 
 | | X-ray structure of the Haloalkane dehalogenase HOB (HaloTag7-based Oligonucleotide Binder) labeled with a chloroalkane-tetramethylrhodamine fluorophore substrate | | Descriptor: | ACETATE ION, CALCIUM ION, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
8B6N
 
 | |
8B6P
 
 | |
3AYK
 
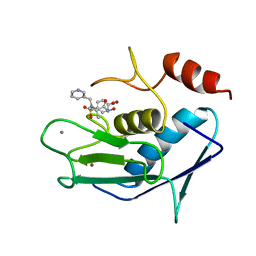 | | CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, PROTEIN (COLLAGENASE), ... | | Authors: | Powers, R, Moy, F.J. | | Deposit date: | 1999-02-01 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound.
Biochemistry, 38, 1999
|
|
3BY7
 
 | |
