4AIS
 
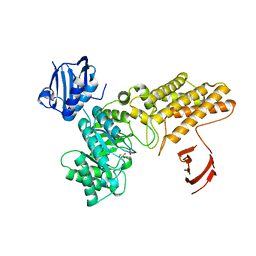 | | A complex structure of BtGH84 | | Descriptor: | GLYCEROL, GLYCOLIC ACID, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2012-02-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism of Vertebrate Amino Sugars with N-Glycolyl Groups: Intracellular Beta-O-Linked N-Glycolylglucosamine (Glcngc), Udp-Glcngc, and the Biochemical and Structural Rationale for the Substrate Tolerance of Beta-O-Linked Beta-N-Acetylglucosaminidase.
J.Biol.Chem., 287, 2012
|
|
4AIU
 
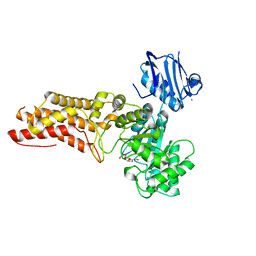 | | A complex structure of BtGH84 | | Descriptor: | (3AR,5R,6S,7R,7AR)-2,5-BIS(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]OXAZOLE-6,7-DIOL, CALCIUM ION, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2012-02-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Metabolism of Vertebrate Amino Sugars with N-Glycolyl Groups: Intracellular Beta-O-Linked N-Glycolylglucosamine (Glcngc), Udp-Glcngc, and the Biochemical and Structural Rationale for the Substrate Tolerance of Beta-O-Linked Beta-N-Acetylglucosaminidase.
J.Biol.Chem., 287, 2012
|
|
2VVN
 
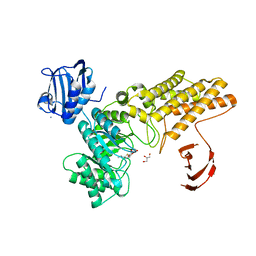 | | BtGH84 in complex with NH-Butylthiazoline | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, AMMONIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Potent Mechanism-Inspired O-Glcnacase Inhibitor that Blocks Phosphorylation of Tau in Vivo.
Nat.Chem.Biol., 4, 2008
|
|
2WCA
 
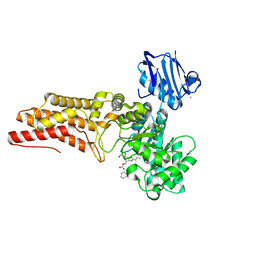 | | BtGH84 in complex with n-butyl pugnac | | Descriptor: | CALCIUM ION, O-GLCNACASE BT_4395, [[(3R,4R,5S,6R)-3-(BUTANOYLAMINO)-4,5-DIHYDROXY-6-(HYDROXYMETHYL)OXAN-2-YLIDENE]AMINO] N-PHENYLCARBAMATE | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into a Strategy for Attenuating Ampc- Mediated Beta-Lactam Resistance: Structural Basis for Selective Inhibition of the Glycoside Hydrolase Nagz.
Protein Sci., 18, 2009
|
|
2XJ7
 
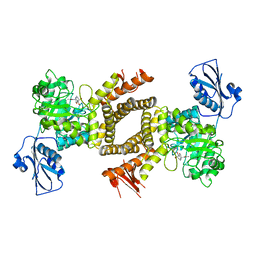 | | BtGH84 in complex with 6-acetamido-6-deoxy-castanospermine | | Descriptor: | 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, CALCIUM ION, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of O-Glcnacase Using a Potent and Cell-Permeable Inhibitor Does not Induce Insulin Resistance in 3T3-L1 Adipocytes.
Chem.Biol., 17, 2010
|
|
8KAE
 
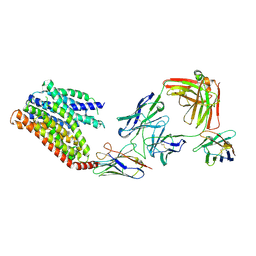 | | 16d-bound human SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, NbFab chain L, NbFab-H chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
2KDM
 
 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | Descriptor: | DESIGNED PROTEIN | | Authors: | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-12-29 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | From the Cover: A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KDL
 
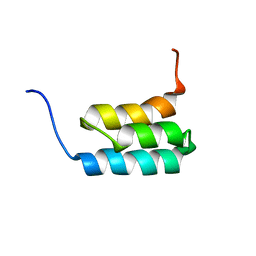 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | Descriptor: | designed protein | | Authors: | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-12-29 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1JSP
 
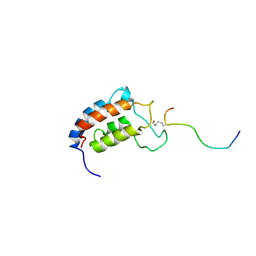 | | NMR Structure of CBP Bromodomain in complex with p53 peptide | | Descriptor: | CREB-BINDING PROTEIN, tumor protein p53 | | Authors: | He, Y, Mujtaba, S, Zeng, L, Yan, S, Zhou, M.-M. | | Deposit date: | 2001-08-17 | | Release date: | 2002-08-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation.
Mol.Cell, 13, 2004
|
|
1JEW
 
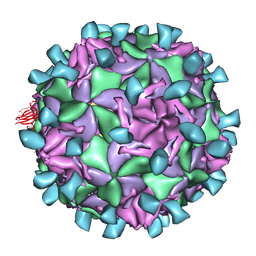 | |
7LSY
 
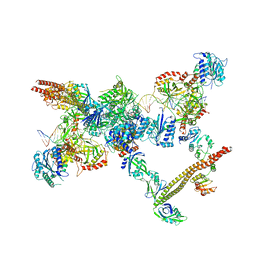 | | NHEJ Short-range synaptic complex | | Descriptor: | DNA (26-MER), DNA (5'-D(P*CP*AP*AP*TP*GP*AP*AP*AP*CP*GP*GP*AP*AP*CP*AP*GP*TP*CP*AP*G)-3'), DNA (5'-D(P*GP*TP*TP*CP*TP*TP*AP*GP*TP*AP*TP*AP*TP*A)-3'), ... | | Authors: | He, Y, Chen, S. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of long-range to short-range synaptic transition in NHEJ.
Nature, 593, 2021
|
|
2MH8
 
 | | GA-79-MBP cs-rosetta structures | | Descriptor: | GA-79-MBP, maltose binding protein | | Authors: | He, Y, Chen, Y, Porter, L, Bryan, P, Orban, J. | | Deposit date: | 2013-11-19 | | Release date: | 2015-04-08 | | Method: | SOLUTION NMR | | Cite: | Subdomain interactions foster the design of two protein pairs with 80% sequence identity but different folds.
Biophys.J., 108, 2015
|
|
8JXT
 
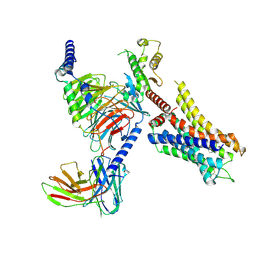 | | Histamine-bound H4R/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
8JXW
 
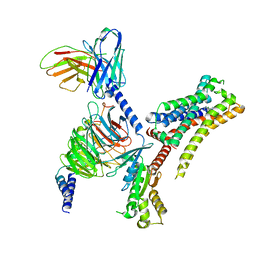 | | VUF6884-bound H4R/Gi complex | | Descriptor: | 2-chloranyl-6-(4-methylpiperazin-1-yl)benzo[b][1,4]benzoxazepine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
8JXX
 
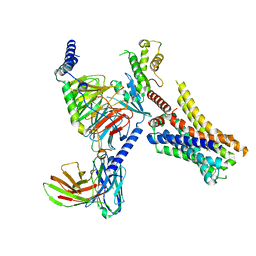 | | Clobenpropit-bound H4R/Gi complex | | Descriptor: | 3-(1~{H}-imidazol-4-yl)propyl ~{N}'-[(4-chlorophenyl)methyl]carbamimidothioate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
8JXV
 
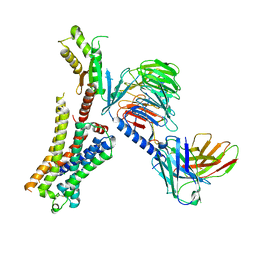 | | Clozapine-bound H4R/Gi complex | | Descriptor: | 3-chloranyl-6-(4-methylpiperazin-1-yl)-11~{H}-benzo[b][1,4]benzodiazepine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
2MJY
 
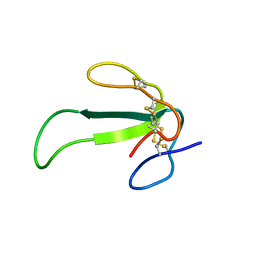 | | Solution structure of synthetic Mamba-1 peptide | | Descriptor: | Mambalgin-1 | | Authors: | He, Y, Wu, F, Tian, C. | | Deposit date: | 2014-01-21 | | Release date: | 2015-01-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | One-pot hydrazide-based native chemical ligation for efficient chemical synthesis and structure determination of toxin Mambalgin-1
Chem.Commun.(Camb.), 50, 2014
|
|
3KX2
 
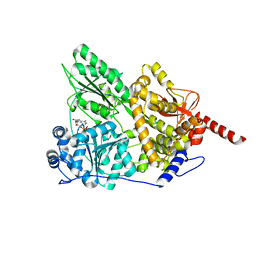 | |
8IYS
 
 | | TUG891-bound FFAR4 in complex with Gq | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4K
 
 | | GW9508-bound FFAR4 in complex with Gq | | Descriptor: | 3-(4-{[(3-phenoxyphenyl)methyl]amino}phenyl)propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4L
 
 | | DHA-bound FFAR4 in complex with Gq | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4I
 
 | | DHA-bound FFAR4 in complex with Gs | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
7YUF
 
 | | apo human SPNS2 | | Descriptor: | NbFab H-chain, NbFab L-chain, Sphingosine-1-phosphate transporter SPNS2, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YUD
 
 | | FTY720p-bound human SPNS2 | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, NbFab L-chain, NbFab-H-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YUB
 
 | | S1P-bound human SPNS2 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, NbFab-H-chain, NbFab-L-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
