9C2H
 
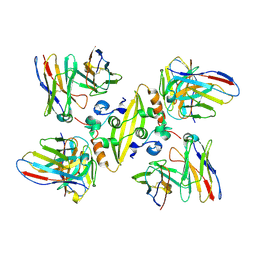 | | SARS-CoV-2 Nucleocapsid Dimerization Domain bound to Fab-NP1E9 and Fab-NP3B4 | | Descriptor: | Antibody Fab NP1-E9 Heavy Chain (variable region), Antibody Fab NP1-E9 Light Chain (variable region), Antibody Fab NP3-B4 Heavy Chain (variable region), ... | | Authors: | Landeras-Bueno, S, Hariharan, C, Diaz Avalos, R, Ollmann Saphire, E. | | Deposit date: | 2024-05-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural stabilization of the intrinsically disordered SARS-CoV-2 N by binding to RNA sequences engineered from the viral genome
To Be Published
|
|
7TN9
 
 | | Structure of the Inmazeb cocktail and resistance to escape against Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, GP2, ... | | Authors: | Rayaprolu, V, Fulton, B, Rafique, A, Arturo, E, Williams, D, Hariharan, C, Callaway, H, Parvate, A, Schendel, S.L, Parekh, D, Hui, S, Shaffer, K, Pascal, K.E, Wloga, E, Giordano, S, Copin, R, Franklin, M, Boytz, R.M, Donahue, C, Davey, R, Baum, A, Kyratsous, C.A, Saphire, E.O. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the Inmazeb cocktail and resistance to Ebola virus escape.
Cell Host Microbe, 31, 2023
|
|
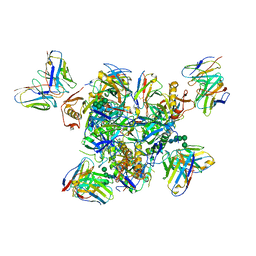
9AT8
 
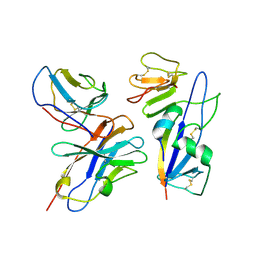 | | Fab 77-stabilized MeV F ectodomain fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, mAB 77 heavy chain, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
8UUP
 
 | | Structure of the Measles virus Fusion protein in the pre-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
8UT2
 
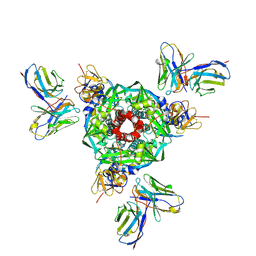 | | Pre-fusion Measles virus fusion protein complexed with Fab 77 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2023-10-30 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
8UUQ
 
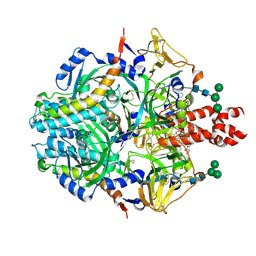 | |
8UTF
 
 | |
8DPM
 
 | |
8DPL
 
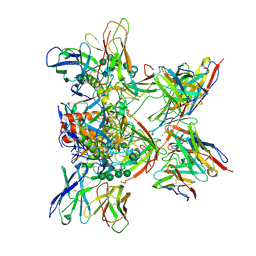 | |
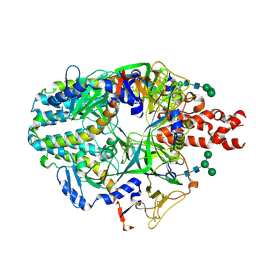
3SUO
 
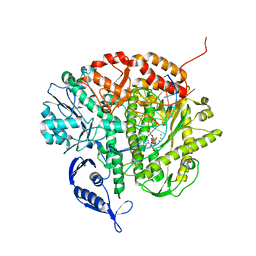 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite 2AP (GC rich sequence) | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SUN
 
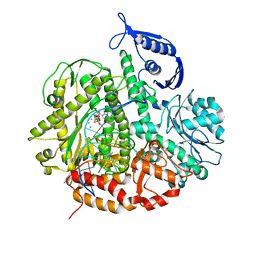 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite 2AP (AT rich sequence) | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SQ2
 
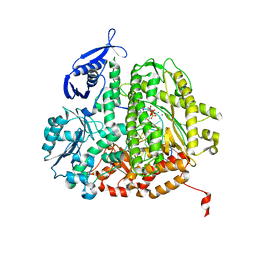 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite 2AP (AT rich sequence) | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SQ4
 
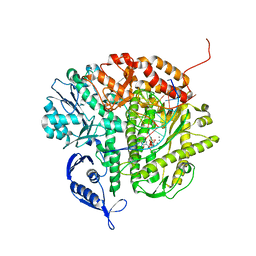 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite 2AP (GC rich sequence) | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SUQ
 
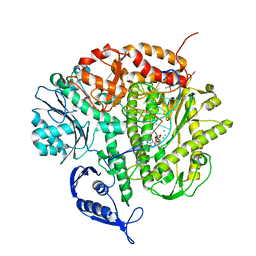 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dCTP Opposite 2AP (AT rich sequence) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SUP
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dCTP Opposite 2AP (GC rich sequence) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
8E1G
 
 | | SARS-CoV-2 RBD in complex with Omicron-neutralizing antibody 2A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2A10 Fab, heavy chain, ... | | Authors: | Wasserman, H, Hastie, K.M, Buck, T.K, Saphire, E.O. | | Deposit date: | 2022-08-10 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
8F0H
 
 | | Structure of SARS-CoV-2 spike with antibody Fabs 2A10 and 1H2 (Local refinement of the RBD and Fabs 1H2 and 2A10) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 1H2 heavy chain, Antibody Fab 1H2 light chain, ... | | Authors: | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
8F0G
 
 | | Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody Fab 1C3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 1C3 Fab Heavy Chain, ... | | Authors: | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
