3IMN
 
 | |
3INA
 
 | | Crystal structure of heparin lyase I H151A mutant complexed with a dodecasaccharide heparin | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, CALCIUM ION, Heparin lyase I | | Authors: | Han, Y.H, Ryu, K.S, Kim, H.Y, Jeon, Y.H. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I
J.Biol.Chem., 284, 2009
|
|
3IN9
 
 | | Crystal structure of heparin lyase I complexed with disaccharide heparin | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparin lyase I | | Authors: | Han, Y.H, Ryu, K.S, Kim, H.Y, Jeon, Y.H. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I
J.Biol.Chem., 284, 2009
|
|
6M1T
 
 | |
3ISO
 
 | | Crystal structure of 26 kDa GST of Clonorchis sinensis in P3221 symmetry | | Descriptor: | GLUTATHIONE, Putative glutathione transferase, SULFATE ION, ... | | Authors: | Han, Y.H, Seo, H.A, Kim, G.H, Chung, Y.J. | | Deposit date: | 2009-08-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A histidine substitution confers metal binding affinity to a Schistosoma japonicum Glutathione S-transferase.
Protein Expr.Purif., 73, 2010
|
|
4L5L
 
 | | Crystal structure of 26 kDa GST of Clonorchis sinensis in P212121 symmetry | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Putative glutathione transferase, ... | | Authors: | Chung, Y.J, Han, Y.H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-09-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of 26kDa Clonorchis sinensis glutathione S-transferase reveal zinc binding and putative metal binding.
Biochem.Biophys.Res.Commun., 438, 2013
|
|
4L5O
 
 | | Crystal structure of 26 kDa GST D26H mutant of Clonorchis sinensis | | Descriptor: | GLUTATHIONE, Putative glutathione transferase, SULFATE ION, ... | | Authors: | Chung, Y.J, Han, Y.H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structures of 26kDa Clonorchis sinensis glutathione S-transferase reveal zinc binding and putative metal binding.
Biochem.Biophys.Res.Commun., 438, 2013
|
|
4WHK
 
 | | A New Class of Peptidomimetics Targeting the Polo-box Domain of Polo-like kinase 1 | | Descriptor: | C6H5(CH2)8-DERIVATIZED PEPTIDE INHIBITOR, Serine/threonine-protein kinase PLK1 | | Authors: | Bang, J.K, Han, Y.H, Ahn, M.J, Lee, K.S. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new class of peptidomimetics targeting the polo-box domain of polo-like kinase 1.
J.Med.Chem., 58, 2015
|
|
4WHL
 
 | | A New Class of Peptidomimetics Targeting the Polo-box Domain of Polo-like kinase 1 | | Descriptor: | C6H5(CH2)8-DERIVATIZED PEPTIDE INHIBITOR, Serine/threonine-protein kinase PLK1 | | Authors: | Bang, J.K, Han, Y.H, Ahn, M.J, Lee, K.S. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A new class of peptidomimetics targeting the polo-box domain of polo-like kinase 1.
J.Med.Chem., 58, 2015
|
|
2EFZ
 
 | |
5DNJ
 
 | | Mouse Polo-box domain and Peptide analog 702 | | Descriptor: | Serine/threonine-protein kinase PLK1, peptide 707-56A-SER-TPO-NH2 | | Authors: | Namgoong, S, Han, Y.H. | | Deposit date: | 2015-09-10 | | Release date: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of Emi2 by Polo-like kinase 1 and development of peptidomimetics blocking oocyte maturation and fertilization.
Sci Rep, 5, 2015
|
|
6J3X
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotriose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-01-06 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
6IYG
 
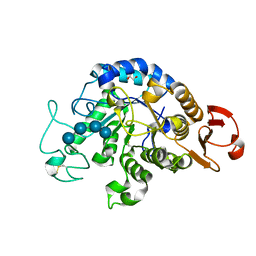 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotetraose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-15 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
4LKX
 
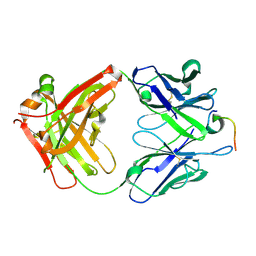 | | Humanized antibody 4B12 Fab complexed with a CemX segment | | Descriptor: | CemX segment, Fab fragment heavy chain, Fab fragment light chain | | Authors: | Chu, H.M, Wright, J, Chan, Y.H, Lin, C.J, Chang, T.W, Lim, C. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Two potential therapeutic antibodies bind to a peptide segment of membrane-bound IgE in different conformations.
Nat Commun, 5, 2014
|
|
8XBF
 
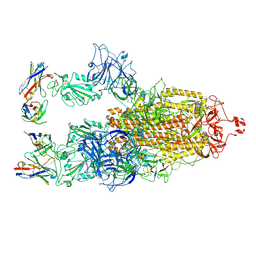 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, O5C2, heavy chain, ... | | Authors: | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|
3IKW
 
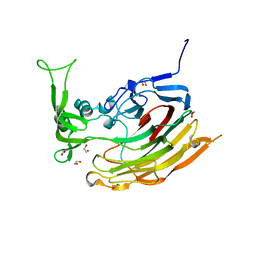 | | Structure of Heparinase I from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Heparin lyase I | | Authors: | Garron, M.L, Cygler, M, Shaya, D. | | Deposit date: | 2009-08-06 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I.
J.Biol.Chem., 284, 2009
|
|
3ILR
 
 | | Structure of Heparinase I from Bacteroides thetaiotaomicron in complex with tetrasaccharide product | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(4-1)-4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid, 4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ... | | Authors: | Garron, M.L, Cygler, M, Shaya, D. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I.
J.Biol.Chem., 284, 2009
|
|
5YGF
 
 | |
5YGD
 
 | |
5YGB
 
 | |
5YGC
 
 | |
6LYI
 
 | |
6LYH
 
 | | Crystal structure of tea N9-methyltransferase CkTcS in complex with SAH and 1,3,7-trimethyluric acid | | Descriptor: | 1,3,7-trimethyl-9H-purine-2,6,8-trione, N-methyltransferase CkTcS, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wang, Y, Zhang, Z.-M. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15000319 Å) | | Cite: | Identification and characterization of N9-methyltransferase involved in converting caffeine into non-stimulatory theacrine in tea.
Nat Commun, 11, 2020
|
|
4WHH
 
 | |
7XHP
 
 | |
