4IGT
 
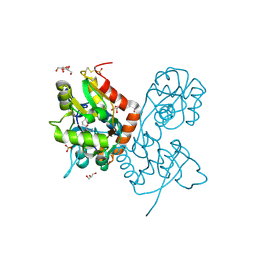 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist ZA302 at 1.24A resolution | | 分子名称: | (4R)-4-{3-[hydroxy(methyl)amino]-3-oxopropyl}-L-glutamic acid, GLYCEROL, Glutamate receptor 2, ... | | 著者 | Larsen, A.P, Venskutonyte, R, Gajhede, M, Kastrup, J.S, Frydenvang, K. | | 登録日 | 2012-12-18 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters.
J.Med.Chem., 56, 2013
|
|
6LYU
 
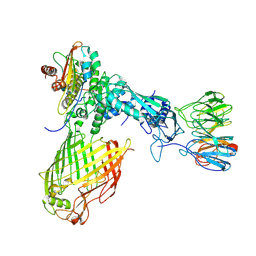 | | Structure of the BAM complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Xiao, L, Huang, Y. | | 登録日 | 2020-02-15 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates.
Faseb J., 35, 2021
|
|
6LYQ
 
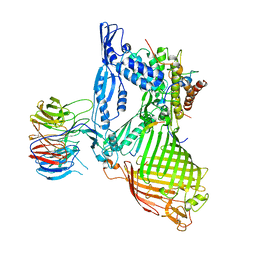 | | Structure of the BAM complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Xiao, L, Huang, Y. | | 登録日 | 2020-02-15 | | 公開日 | 2021-01-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates.
Faseb J., 35, 2021
|
|
6LYS
 
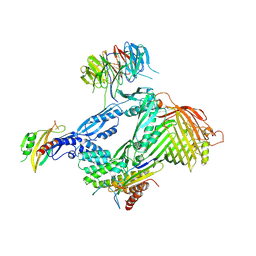 | | Structure of the BAM complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Xiao, L, Huang, Y. | | 登録日 | 2020-02-15 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates.
Faseb J., 35, 2021
|
|
6F29
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2-Amino-2-carboxyethyl]-5,7-dihydrothieno[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.6A | | 分子名称: | (2~{S})-2-azanyl-3-[2,4-bis(oxidanylidene)-5,7-dihydrothieno[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2017-11-23 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
6F28
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2'-Amino-2'-carboxyethyl]-6-methyl-5,7-dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.4A | | 分子名称: | (2~{S})-2-azanyl-3-[6-methyl-2,4-bis(oxidanylidene)-5,7-dihydropyrrolo[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2017-11-23 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
5XMZ
 
 | | Verticillium effector PevD1 | | 分子名称: | CALCIUM ION, CHLORIDE ION, Effector protein PevD1 | | 著者 | Liu, X, Zhou, R. | | 登録日 | 2017-05-17 | | 公開日 | 2017-07-05 | | 最終更新日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The asparagine-rich protein NRP interacts with the Verticillium effector PevD1 and regulates the subcellular localization of cryptochrome 2
J. Exp. Bot., 68, 2017
|
|
7N32
 
 | |
7MWG
 
 | | 16-nm repeat microtubule doublet | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Rao, Q, Zhang, K. | | 登録日 | 2021-05-17 | | 公開日 | 2021-09-29 | | 最終更新日 | 2021-10-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures of outer-arm dynein array on microtubule doublet reveal a motor coordination mechanism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4PT1
 
 | |
7N9Z
 
 | | E. coli cytochrome bo3 in MSP nanodisc | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | 著者 | Vallese, F, Clarke, O.B. | | 登録日 | 2021-06-19 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (2.19 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo 3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4QF9
 
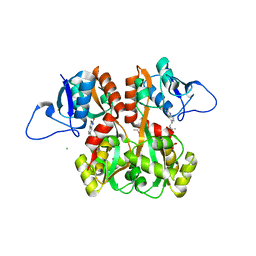 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with (S)-2-amino-4-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-6-yl)butanoic acid at 2.28 A resolution | | 分子名称: | (2S)-2-amino-4-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-6-yl)butanoic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Kristensen, C.M, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2014-05-20 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Binding Mode of an alpha-Amino Acid-Linked Quinoxaline-2,3-dione Analogue at Glutamate Receptor Subtype GluK1.
ACS Chem Neurosci, 6, 2015
|
|
8GQ6
 
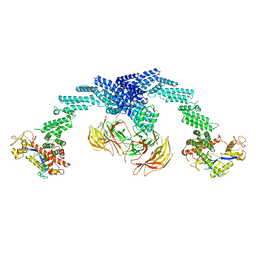 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | 登録日 | 2022-08-29 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H38
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.52 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H36
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H35
 
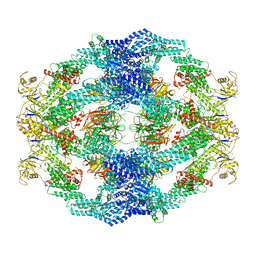 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 octameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.41 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3R
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8 dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.36 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H34
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.99 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.51 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H33
 
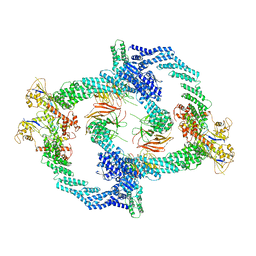 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 tetrameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.86 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3Q
 
 | | Cryo-EM Structure of the CAND1-Cul3-Rbx1 complex | | 分子名称: | Cullin-3, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.73 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5YMV
 
 | | Crystal structure of 9-mer peptide from influenza virus in complex with BF2*1201 | | 分子名称: | ALA-VAL-LYS-GLY-VAL-GLY-THR-MET-VAL, Beta-2-microglobulin, Class I histocompatibility antigen, ... | | 著者 | Xiao, J, Xiang, W, Qi, J, Chai, Y, Liu, W.J, Gao, G.F. | | 登録日 | 2017-10-22 | | 公開日 | 2018-10-24 | | 最終更新日 | 2019-07-31 | | 実験手法 | X-RAY DIFFRACTION (2.197 Å) | | 主引用文献 | An Invariant Arginine in Common with MHC Class II Allows Extension at the C-Terminal End of Peptides Bound to Chicken MHC Class I.
J Immunol., 201, 2018
|
|
5YMW
 
 | | Crystal structure of 8-mer peptide from Rous sarcoma virus in complex with BF2*1201 | | 分子名称: | Beta-2-microglobulin, Class I histocompatibility antigen, F10 alpha chain, ... | | 著者 | Xiao, J, Xiang, W, Qi, J, Chai, Y, Liu, W.J, Gao, G.F. | | 登録日 | 2017-10-22 | | 公開日 | 2018-10-24 | | 最終更新日 | 2019-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | An Invariant Arginine in Common with MHC Class II Allows Extension at the C-Terminal End of Peptides Bound to Chicken MHC Class I.
J Immunol., 201, 2018
|
|
