6OK3
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6ONW
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6ORC
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6ORK
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
5H3D
 
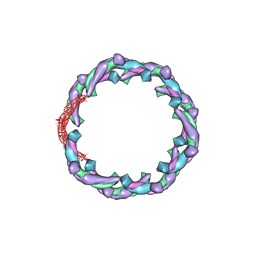 | | Helical structure of membrane tubules decorated by ACAP1 (BARPH doamin) protein by cryo-electron microscopy and MD simulation | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | Authors: | Chan, C, Pang, X.Y, Zhang, Y, Sun, F, Fan, J. | | Deposit date: | 2016-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | ACAP1 assembles into an unusual protein lattice for membrane deformation through multiple stages.
Plos Comput.Biol., 15, 2019
|
|
4W9R
 
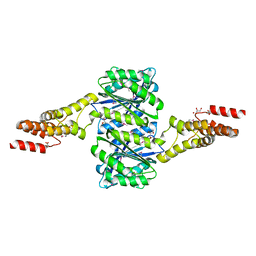 | | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271 | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271
To Be Published
|
|
4E2G
 
 | | Crystal structure of Cupin fold protein Sthe2323 from Sphaerobacter thermophilus | | Descriptor: | ACETATE ION, Cupin 2 conserved barrel domain protein, NICKEL (II) ION, ... | | Authors: | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-03-08 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of Cupin fold protein Sthe2323 from Sphaerobacter thermophilus
To be Published
|
|
4EMY
 
 | |
8GH5
 
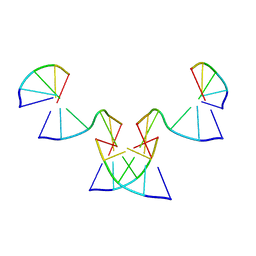 | | Implementing Logic Gates in DNA Crystal Engineering | | Descriptor: | DNA (5'-D(*AP*GP*AP*CP*G)-3'), DNA (5'-D(*CP*TP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), ... | | Authors: | Zhang, C, Paluzzi, V.E, Sha, R, Jonoska, N, Mao, C. | | Deposit date: | 2023-03-09 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Implementing Logic Gates by DNA Crystal Engineering.
Adv Mater, 35, 2023
|
|
3F9U
 
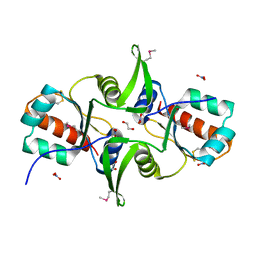 | | Crystal structure of C-terminal domain of putative exported cytochrome C biogenesis-related protein from Bacteroides fragilis | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Putative exported cytochrome C biogenesis-related protein | | Authors: | Chang, C, Tesar, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of C-terminal domain of putative exported cytochrome C biogenesis-related protein from Bacteroides fragilis
To be Published
|
|
7M92
 
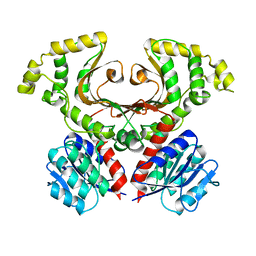 | |
7MH7
 
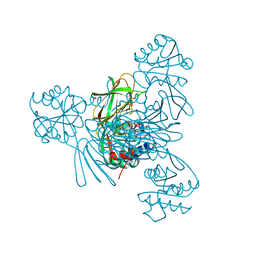 | |
1Z7A
 
 | | Crystal structure of probable Polysaccharide deacetylase from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-24 | | Release date: | 2005-05-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of probable Polysaccharide deacetylase from Pseudomonas aeruginosa PAO1
To be Published
|
|
3EYT
 
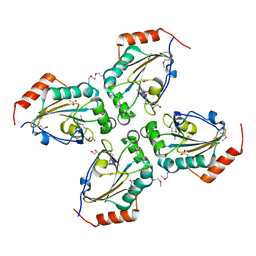 | | Crystal structure of Thioredoxin-like superfamily protein SPOA0173 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, uncharacterized protein SPOA0173 | | Authors: | Chang, C, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Thioredoxin-like superfamily protein SPOA0173
To be Published
|
|
1Z0X
 
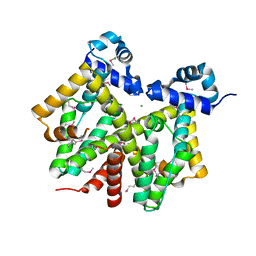 | | Crystal structure of transcriptional regulator, tetR Family from Enterococcus faecalis V583 | | Descriptor: | CHLORIDE ION, transcriptional regulator, TetR family | | Authors: | Chang, C, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-02 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of transcriptional regulator, tetR Family from Enterococcus faecalis V583
To be Published
|
|
3FH0
 
 | | Crystal structure of putative universal stress protein KPN_01444 - ATPase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, putative universal stress protein KPN_01444 | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2008-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative universal stress protein KPN_01444 - ATPase
To be Published
|
|
3F6V
 
 | | Crystal structure of Possible transcriptional regulator for arsenical resistance | | Descriptor: | MAGNESIUM ION, Possible transcriptional regulator, ArsR family protein | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Possible transcriptional regulator for arsenical resistance from Rhodococcus sp.
To be Published
|
|
4MTN
 
 | |
5GP1
 
 | | Crystal structure of ZIKV NS5 Methyltransferase in complex with GTP and SAH | | Descriptor: | NICKEL (II) ION, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-directed RNA polymerase NS5, ... | | Authors: | Zhang, C, Jin, T. | | Deposit date: | 2016-07-30 | | Release date: | 2016-12-07 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design
Biochem. Biophys. Res. Commun., 492, 2017
|
|
4MJD
 
 | | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747 | | Descriptor: | Ketosteroid isomerase fold protein Hmuk_0747, MAGNESIUM ION, SODIUM ION | | Authors: | Chang, C, Holowicki, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747
To be Published
|
|
4MOZ
 
 | |
6UVZ
 
 | | Amidohydrolase 2 from Bifidobacterium longum subsp. infantis | | Descriptor: | Amidohydrolase 2, CITRIC ACID, NONAETHYLENE GLYCOL | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-11-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Amidohydrolase 2 from Bifidobacterium longum subsp. infantis
To Be Published
|
|
6UX3
 
 | | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae | | Descriptor: | Acetoin dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Chang, C, Skarina, T, Mesa, N, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-06 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae
To Be Published
|
|
3K67
 
 | | Crystal structure of protein af1124 from archaeoglobus fulgidus | | Descriptor: | PHOSPHATE ION, putative dehydratase AF1124 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Protein Af1124 from Archaeoglobus Fulgidus
To be Published
|
|
3K6A
 
 | |
