3TS8
 
 | |
7P2G
 
 | |
1T4W
 
 | | Structural Differences in the DNA Binding Domains of Human p53 and its C. elegans Ortholog Cep-1: Structure of C. elegans Cep-1 | | Descriptor: | C.Elegans p53 tumor suppressor-like transcription factor, ZINC ION | | Authors: | Huyen, Y, Jeffrey, P.D, Derry, W.B, Rothman, J.H, Pavletich, N.P, Stavridi, E.S, Halazonetis, T.D. | | Deposit date: | 2004-04-30 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Differences in the DNA Binding Domains of Human p53 and Its C. elegans Ortholog Cep-1.
Structure, 12, 2004
|
|
1XNI
 
 | | Tandem Tudor Domain of 53BP1 | | Descriptor: | Tumor suppressor p53-binding protein 1 | | Authors: | Huyen, Y, Zgheib, O, DiTullio Jr, R.A, Gorgoulis, V.G, Zacharatos, P, Petty, T.J, Sheston, E.A, Mellert, H.S, Stavridi, E.S, Halazonetis, T.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks
Nature, 432, 2004
|
|
1LGP
 
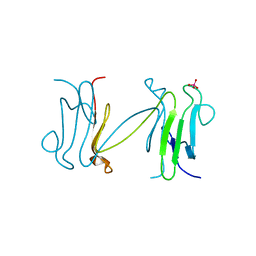 | | Crystal structure of the FHA domain of the Chfr mitotic checkpoint protein complexed with tungstate | | Descriptor: | TUNGSTATE(VI)ION, cell cycle checkpoint protein CHFR | | Authors: | Stavridi, E.S, Huyen, Y, Loreto, I.R, Scolnick, D.M, Halazonetis, T.D, Pavletich, N.P, Jeffrey, P.D. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the FHA domain of the Chfr mitotic checkpoint protein and its complex with tungstate.
Structure, 10, 2002
|
|
1LGQ
 
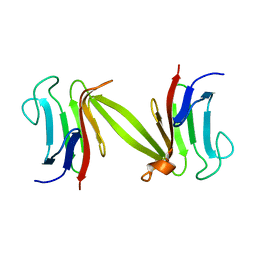 | | Crystal structure of the FHA domain of the Chfr mitotic checkpoint protein | | Descriptor: | cell cycle checkpoint protein CHFR | | Authors: | Stavridi, E.S, Huyen, Y, Loreto, I.R, Scolnick, D.M, Halazonetis, T.D, Pavletich, N.P, Jeffrey, P.D. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the FHA domain of the Chfr mitotic checkpoint protein and its complex with tungstate.
Structure, 10, 2002
|
|
4MZI
 
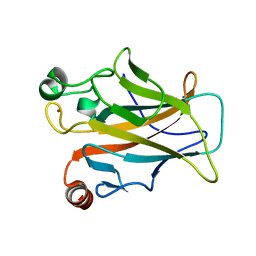 | | Crystal structure of a human mutant p53 | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Emamzadah, S, Tropia, L, Vincenti, I, Falquet, B, Halazonetis, T.D. | | Deposit date: | 2013-09-30 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Reversal of the DNA-Binding-Induced Loop L1 Conformational Switch in an Engineered Human p53 Protein.
J.Mol.Biol., 426, 2014
|
|
4MZR
 
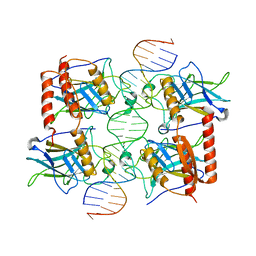 | | Crystal structure of a polypeptide p53 mutant bound to DNA | | Descriptor: | Cellular tumor antigen p53, ZINC ION, consensus DNA anti-sense strand, ... | | Authors: | Emamzadah, S.T, Tropia, L, Vincenti, I, Falquet, B, Halazonetis, T.D. | | Deposit date: | 2013-09-30 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Reversal of the DNA-Binding-Induced Loop L1 Conformational Switch in an Engineered Human p53 Protein.
J.Mol.Biol., 426, 2014
|
|
3Q06
 
 | |
3Q01
 
 | |
3Q05
 
 | |
3VEM
 
 | | Structural basis of transcriptional gene silencing mediated by Arabidopsis MOM1 | | Descriptor: | Helicase protein MOM1 | | Authors: | Nishikura, T, Petty, T.J, Halazonetis, T, Paszkowski, J, Thore, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Transcriptional Gene Silencing Mediated by Arabidopsis MOM1.
PLOS GENET., 8, 2012
|
|
1A1U
 
 | | SOLUTION STRUCTURE DETERMINATION OF A P53 MUTANT DIMERIZATION DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | P53 | | Authors: | Mccoy, M.A, Stavridi, E.S, Waterman, J.L.F, Wieczorek, A, Opella, S.J, Halezonetis, T.D. | | Deposit date: | 1997-12-16 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Hydrophobic side-chain size is a determinant of the three-dimensional structure of the p53 oligomerization domain.
EMBO J., 16, 1997
|
|
4QLI
 
 | |
