2LXG
 
 | |
2M62
 
 | |
2LO9
 
 | | NMR solution structure of Mu-contoxin BuIIIB | | Descriptor: | Mu-conotoxin BuIIIB | | Authors: | Kuang, Z. | | Deposit date: | 2012-01-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mammalian neuronal sodium channel blocker mu-conotoxin BuIIIB has a structured N-terminus that influences potency.
Acs Chem.Biol., 8, 2013
|
|
2LOC
 
 | | Conotoxin analogue [D-Ala2]BuIIIB | | Descriptor: | Mu-conotoxin BuIIIB | | Authors: | Kuang, Z. | | Deposit date: | 2012-01-21 | | Release date: | 2013-01-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mammalian neuronal sodium channel blocker mu-conotoxin BuIIIB has a structured N-terminus that influences potency.
Acs Chem.Biol., 8, 2013
|
|
6F3T
 
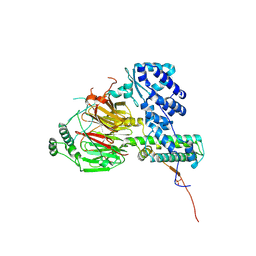 | | Crystal structure of the human TAF5-TAF6-TAF9 complex | | Descriptor: | CHLORIDE ION, Transcription initiation factor TFIID subunit 5, Transcription initiation factor TFIID subunit 6, ... | | Authors: | Haffke, M, Berger, I. | | Deposit date: | 2017-11-28 | | Release date: | 2018-12-05 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chaperonin CCT checkpoint function in basal transcription factor TFIID assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6N1K
 
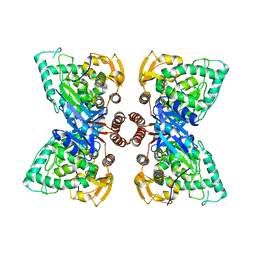 | |
5YJE
 
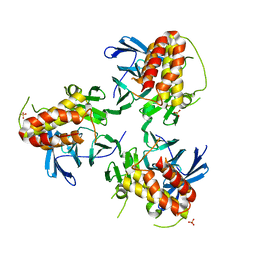 | | Crystal structure of HIRA(644-1017) | | Descriptor: | Protein HIRA, SULFATE ION | | Authors: | Sato, Y, Senda, M, Senda, T. | | Deposit date: | 2017-10-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit
Nat Commun, 9, 2018
|
|
5DEN
 
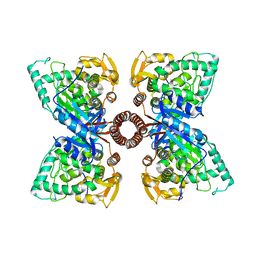 | |
7LQ5
 
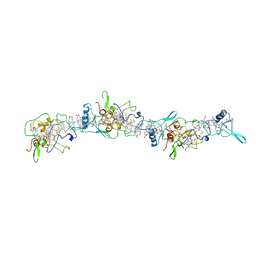 | | Cryo-EM structure of OmcZ nanowire from Geobacter sulfurreducens | | Descriptor: | Cytochrome c, HEME C | | Authors: | Gu, Y, Srikanth, V, Malvankar, N.S, Samatey, F.A. | | Deposit date: | 2021-02-13 | | Release date: | 2022-08-24 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of Geobacter cytochrome OmcZ identifies mechanism of nanowire assembly and conductivity.
Nat Microbiol, 8, 2023
|
|
7N9H
 
 | |
7LZA
 
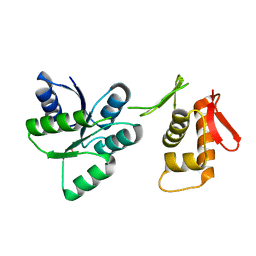 | | Activated form of VanR from S. coelicolor | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Putative two-component system response regulator | | Authors: | Maciunas, L.J, Loll, P.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of full-length VanR from Streptomyces coelicolor in both the inactive and activated states.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7LZ9
 
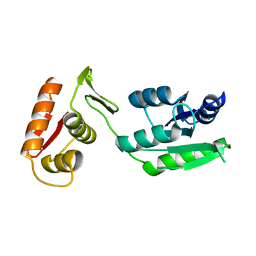 | | Inactive form of VanR from S. coelicolor | | Descriptor: | MAGNESIUM ION, Putative two-component system response regulator | | Authors: | Maciunas, L.J, Loll, P.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of full-length VanR from Streptomyces coelicolor in both the inactive and activated states.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
