5UPE
 
 | |
8IKL
 
 | | Cryo-EM structure of the CD97-G13 complex | | Descriptor: | Adhesion G protein-coupled receptor E5, Guanine nucleotide-binding protein G(13) subunit alpha isoforms short, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
8GUE
 
 | | Crystal Structure of narbomycin-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450 monooxygenase PikC, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-09-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids.
Nat Commun, 14, 2023
|
|
7F3U
 
 | | Cryo-EM structure of human TMEM120A in the CoASH-free state | | Descriptor: | Transmembrane protein 120A | | Authors: | Rong, Y, Gao, Y.W, Song, D.F, Zhao, Y, Liu, Z.F. | | Deposit date: | 2021-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | TMEM120A contains a specific coenzyme A-binding site and might not mediate poking- or stretch-induced channel activities in cells.
Elife, 10, 2021
|
|
7F3T
 
 | |
7CBB
 
 | |
7D60
 
 | | Cryo-EM Structure of human CALHM5 in the presence of rubidium red | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7D65
 
 | | Cryo-EM Structure of human CALHM5 in the presence of Ca2+ | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7D61
 
 | | Cryo-EM Structure of human CALHM5 in the presence of EDTA | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | Descriptor: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVF
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH062 | | Descriptor: | 2-(4-azanylpiperidin-1-yl)-6-chloranyl-N-(1-methylimidazol-4-yl)quinazolin-4-amine, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5YAD
 
 | | Crystal structure of Marf1 Lotus domain from Mus musculus | | Descriptor: | GLYCEROL, Meiosis regulator and mRNA stability factor 1, SULFATE ION | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YAA
 
 | | Crystal structure of Marf1 NYN domain from Mus musculus | | Descriptor: | GLYCEROL, Meiosis regulator and mRNA stability factor 1 | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5HMS
 
 | | X-ray structure of human recombinant 5-aminolaevulinic acid dehydratase (hrALAD). | | Descriptor: | Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Butler, D, Erskine, P.T, Cooper, J.B, Shoolingin-Jordan, P.M. | | Deposit date: | 2016-01-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5HNR
 
 | | The X-ray structure of octameric human native 5-aminolaevulinic acid dehydratase. | | Descriptor: | DELTA-AMINO VALERIC ACID, Delta-aminolevulinic acid dehydratase, SULFATE ION, ... | | Authors: | Mills-Davies, N.L, Thompson, D, Shoolingin-Jordan, P.M, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-01-18 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3DX3
 
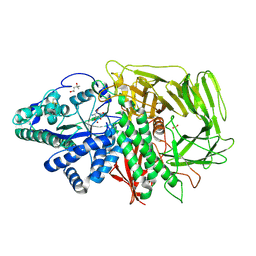 | | Golgi alpha-Mannosidase II in complex with Mannostatin analog (1R,2R,3S,4R,5R)-5-aminocyclopentane-1,2,3,4-tetraol | | Descriptor: | (1R,2R,3S,4R,5R)-5-aminocyclopentane-1,2,3,4-tetrol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-07-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The molecular basis of inhibition of Golgi alpha-mannosidase II by mannostatin A.
Chembiochem, 10, 2009
|
|
3DX0
 
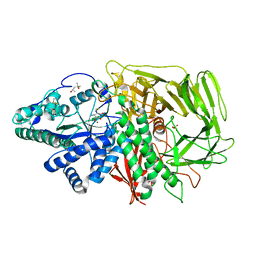 | | Golgi alpha-Mannosidase II in complex with Mannostatin A at pH 5.75 | | Descriptor: | (1R,2R,3R,4S,5R)-4-AMINO-5-(METHYLTHIO)CYCLOPENTANE-1,2,3-TRIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-07-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis of inhibition of Golgi alpha-mannosidase II by mannostatin A.
Chembiochem, 10, 2009
|
|
3DX4
 
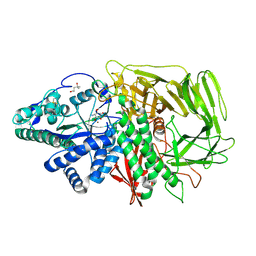 | | Golgi alpha-Mannosidase II in complex with Mannostatin analog (1R,2R,3R,4S,5R)-4-amino-5-methoxycyclopentane-1,2,3-triol | | Descriptor: | (1R,2R,3R,4S,5R)-4-amino-5-methoxycyclopentane-1,2,3-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-07-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The molecular basis of inhibition of Golgi alpha-mannosidase II by mannostatin A.
Chembiochem, 10, 2009
|
|
3DX1
 
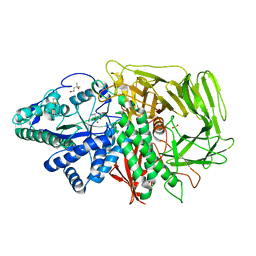 | | Golgi alpha-Mannosidase II in complex with Mannostatin analog (1S,2S,3R,4R)-4-aminocyclopentane-1,2,3-triol | | Descriptor: | (1S,2S,3R,4R)-4-aminocyclopentane-1,2,3-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-07-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The molecular basis of inhibition of Golgi alpha-mannosidase II by mannostatin A.
Chembiochem, 10, 2009
|
|
6LQE
 
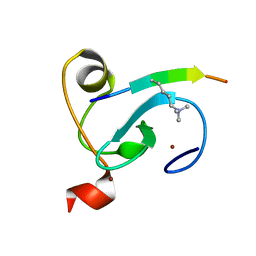 | |
6LQF
 
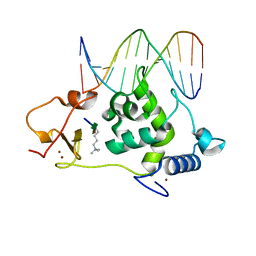 | |
7SIU
 
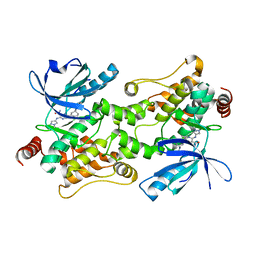 | | Crystal structure of HPK1 (MAP4K1) complex with inhibitor A-745 | | Descriptor: | 6-[(5R)-5-benzamidocyclohex-1-en-1-yl]-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Longenecker, K.L, Korepanova, A, Qiu, W. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | The HPK1 Inhibitor A-745 Verifies the Potential of Modulating T Cell Kinase Signaling for Immunotherapy.
Acs Chem.Biol., 17, 2022
|
|
7T6T
 
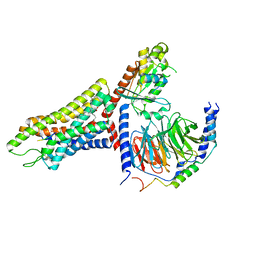 | | Structure of the human FPR1-Gi complex with fMLFII | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6V
 
 | | Structure of the human FPR2-Gi complex with fMLFII | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
