4DG9
 
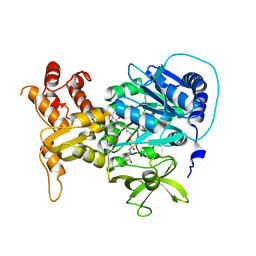 | | Structure of holo-PA1221, an NRPS protein containing adenylation and PCP domains bound to vinylsulfonamide inhibitor | | Descriptor: | 5'-({[(2R,3R)-3-amino-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-{[oxido(oxo)phosphonio]oxy}butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-4-methylpentyl]sulfonyl}amino)-5'-deoxyadenosine, PA1221 | | Authors: | Mitchell, C.A, Shi, C, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of PA1221, a Nonribosomal Peptide Synthetase Containing Adenylation and Peptidyl Carrier Protein Domains.
Biochemistry, 51, 2012
|
|
4G35
 
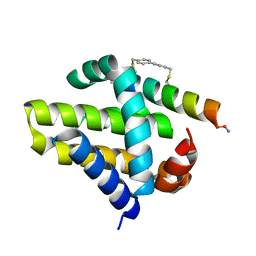 | | Mcl-1 in complex with a biphenyl cross-linked Noxa peptide. | | Descriptor: | 4,4'-bis(bromomethyl)biphenyl, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa BH3 peptide (cysteine-mediated cross-linked) | | Authors: | Drake, E, Edwardraja, S, Lin, Q, Gulick, A.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-12-05 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of proteolytically stable, cell-permeable peptide-based selective Mcl-1 inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
4G36
 
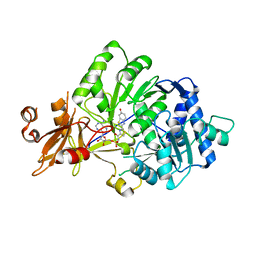 | |
4G37
 
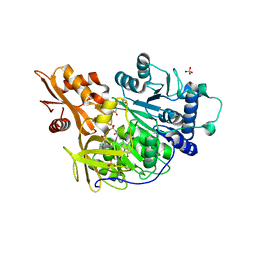 | | Structure of cross-linked firefly luciferase in second catalytic conformation | | Descriptor: | 4,4'-(ethylenediimino)bis[4-oxobutyrate], 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, ... | | Authors: | Sundlov, J.A, Branchini, B.R, Gulick, A.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Crystal structure of firefly luciferase in a second catalytic conformation supports a domain alternation mechanism.
Biochemistry, 51, 2012
|
|
4HKG
 
 | | Crystal Structure of Free-Standing Peptidyl Carrier Protein from Uncharacterized Acinetobacter baumannii Secondary Metabolic Pathway | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Allen, C.L, Gulick, A.M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3002 Å) | | Cite: | Structural and bioinformatic characterization of an Acinetobacter baumannii type II carrier protein.
Acta Crystallogr. D Biol. Crystallogr., 70, 2014
|
|
4JNJ
 
 | | Structure based engineering of streptavidin monomer with a reduced biotin dissociation rate | | Descriptor: | BIOTIN, Streptavidin/Rhizavidin Hybrid, ZINC ION | | Authors: | DeMonte, D, Drake, E.J, Hong Lim, K, Gulick, A.M, Park, S. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structure-based engineering of streptavidin monomer with a reduced biotin dissociation rate.
Proteins, 81, 2013
|
|
4K2G
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A33442372 | | Descriptor: | (2S)-(4-fluorophenyl)[6-(trifluoromethyl)pyridin-2-yl]ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
4K2F
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A08522488 | | Descriptor: | (2S)-(4-chlorophenyl)(6-chloropyridin-2-yl)ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
6VHT
 
 | |
6VHU
 
 | | Klebsiella oxytoca NpsA N-terminal subdomain in space group P21 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Kreitler, D.F, Gulick, A.M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic PathogenKlebsiella oxytoca.
Acs Infect Dis., 6, 2020
|
|
6VHX
 
 | | Klebsiella oxytoca NpsA N-terminal subdomain in complex with 3-hydroxyanthranilyl-AMSN | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-{[(2-amino-3-hydroxybenzene-1-carbonyl)sulfamoyl]amino}-5'-deoxyadenosine, BROMIDE ION, ... | | Authors: | Kreitler, D.F, Gulick, A.M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic PathogenKlebsiella oxytoca.
Acs Infect Dis., 6, 2020
|
|
6VHW
 
 | | Klebsiella oxytoca NpsA N-terminal subdomain in complex with 3-hydroxybenzoyl-AMSN | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-deoxy-5'-{[(3-hydroxybenzene-1-carbonyl)sulfamoyl]amino}adenosine, BROMIDE ION, ... | | Authors: | Kreitler, D.F, Gulick, A.M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic PathogenKlebsiella oxytoca.
Acs Infect Dis., 6, 2020
|
|
6VHZ
 
 | | Klebsiella oxytoca NpsA N-terminal subdomain in complex with anthranilyl-AMSN | | Descriptor: | 5'-{[(benzenecarbonyl)sulfamoyl]amino}-5'-deoxyadenosine, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Kreitler, D.F, Gulick, A.M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic PathogenKlebsiella oxytoca.
Acs Infect Dis., 6, 2020
|
|
6VHV
 
 | | Klebsiella oxytoca NpsA in complex with 3-hydroxyanthranilyl-AMSN | | Descriptor: | 5'-{[(2-amino-3-hydroxybenzene-1-carbonyl)sulfamoyl]amino}-5'-deoxyadenosine, NpsA Adenylation Domain | | Authors: | Kreitler, D.F, Gulick, A.M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic PathogenKlebsiella oxytoca.
Acs Infect Dis., 6, 2020
|
|
6VHY
 
 | | NpsA-ThdA, an artificially fused Adenylation-PCP di-domain NRPS from Klebsiella oxytoca | | Descriptor: | 5'-deoxy-5'-({[(2R)-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-2-(3-hydroxyphenyl)ethyl]sulfonyl}amino)adenosine, NpsA Adenylation Domain, Non-ribosomal peptide synthetase fusion protein | | Authors: | Kreitler, D.F, Gulick, A.M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic PathogenKlebsiella oxytoca.
Acs Infect Dis., 6, 2020
|
|
5JM7
 
 | |
5JM8
 
 | |
5JY4
 
 | | A high magnesium structure of the isochorismate synthase, EntC | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, Isochorismate synthase EntC, MAGNESIUM ION | | Authors: | Meneely, K.M, Sundlov, J.A, Gulick, A.M, Lamb, A.L. | | Deposit date: | 2016-05-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
5JY8
 
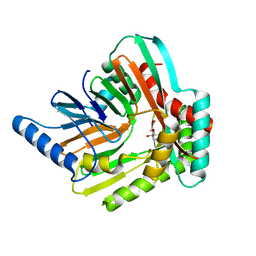 | | An iron-bound structure of the isochorismate synthase EntC | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, FE (III) ION, Isochorismate synthase EntC | | Authors: | Meneely, K.M, Sundlov, J.A, Gulick, A.M, Lamb, A.L. | | Deposit date: | 2016-05-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
5JXZ
 
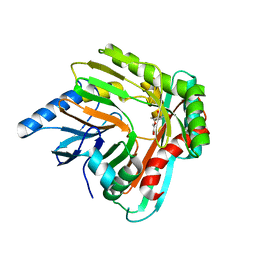 | | A low magnesium structure of the isochorismate synthase, EntC | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, Isochorismate synthase EntC, ... | | Authors: | Meneely, K.M, Sundlov, J.A, Gulick, A.M, Lamb, A.L. | | Deposit date: | 2016-05-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
3O84
 
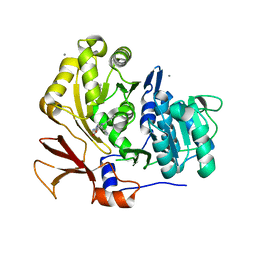 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3O82
 
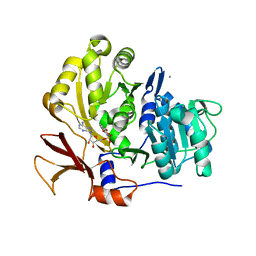 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 5'-O-[N-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine | | Descriptor: | 5'-O-{[(2,3-dihydroxyphenyl)carbonyl]sulfamoyl}adenosine, CALCIUM ION, Peptide arylation enzyme | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3RG2
 
 | |
5I3F
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3H
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, POTASSIUM ION, Triosephosphate isomerase, ... | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
