5O33
 
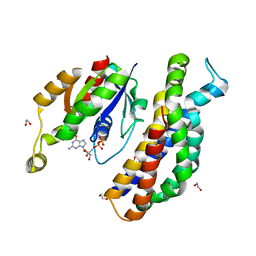 | | A structure of the GEF Kalirin DH1 domain in complex with the small GTPase Rac1 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Kalirin, ... | | Authors: | Gray, J, Krojer, T, Talon, R, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P, von Delft, F. | | Deposit date: | 2017-05-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A structure of the GEF Kalirin DH1 domain in complex with the small GTPase Rac1
To Be Published
|
|
2XJI
 
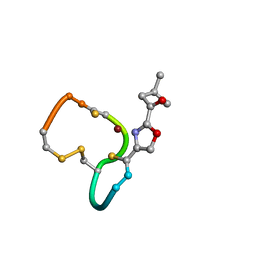 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
2YGI
 
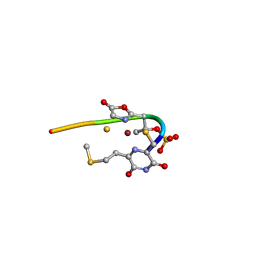 | | Methanobactin HM1 | | Descriptor: | COPPER (II) ION, METHANOBACTIN HM1 | | Authors: | Ghazouani, A, Basle, A, Firbank, S.J, Gray, J, Dennison, C. | | Deposit date: | 2011-04-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Variations in Methanobactin Structure Influences Copper Utilization by Methane-Oxidizing Bacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YGJ
 
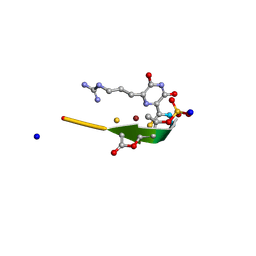 | | Methanobactin MB4 | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB4, SODIUM ION | | Authors: | Ghazouani, A, Basle, A, Firbank, S.J, Gray, J, Dennison, C. | | Deposit date: | 2011-04-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Variations in Methanobactin Structure Influences Copper Utilization by Methane-Oxidizing Bacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2VQA
 
 | | Protein-folding location can regulate Mn versus Cu- or Zn-binding. Crystal Structure of MncA. | | Descriptor: | ACETATE ION, MANGANESE (II) ION, SLL1358 PROTEIN | | Authors: | Tottey, S, Waldron, K.J, Firbank, S.J, Reale, B, Bessant, C, Sato, K, Gray, J, Banfield, M.J, Dennison, C, Robinson, N.J. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Protein-Folding Location Can Regulate Manganese-Binding Versus Copper- or Zinc-Binding.
Nature, 455, 2008
|
|
2XJH
 
 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B, SODIUM ION | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
6FAE
 
 | | The Sec7 domain of IQSEC2 (Brag1) in complex with the small GTPase Arf1 | | Descriptor: | 1,2-ETHANEDIOL, ADP-ribosylation factor 1, IQ motif and SEC7 domain-containing protein 2 | | Authors: | Gray, J, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P, von Delft, F. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Targeting the Small GTPase Superfamily through their Regulatory Proteins.
Angew.Chem.Int.Ed.Engl., 2019
|
|
5LA0
 
 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, SULFATE ION, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, A. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
8EV4
 
 | | NlpC B3 - Trichomonas Vaginalis | | Descriptor: | Clan CA, family C40, NlpC/P60 superfamily cysteine peptidase | | Authors: | Barnett, M.J, Goldstone, D.C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | NlpC/P60 peptidoglycan hydrolases of Trichomonas vaginalis have complementary activities that empower the protozoan to control host-protective lactobacilli.
Plos Pathog., 19, 2023
|
|
8EV5
 
 | | NlpC B3 covalently bound with E64 inhibitor fragment | | Descriptor: | Clan CA, family C40, NlpC/P60 superfamily cysteine peptidase, ... | | Authors: | Barnett, M.J, Goldstone, D.C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | NlpC/P60 peptidoglycan hydrolases of Trichomonas vaginalis have complementary activities that empower the protozoan to control host-protective lactobacilli.
Plos Pathog., 19, 2023
|
|
5MQO
 
 | | Glycoside hydrolase BT_1003 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MSY
 
 | | Glycoside hydrolase BT_1012 | | Descriptor: | AMMONIA, Glycoside hydrolase, PHOSPHATE ION | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQS
 
 | | Sialidase BT_1020 | | Descriptor: | Beta-L-arabinobiosidase, CALCIUM ION, SODIUM ION, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQR
 
 | | Sialidase BT_1020 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-L-arabinobiosidase, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MT2
 
 | | Glycoside hydrolase BT_0996 | | Descriptor: | Beta-galactosidase, GLYCEROL | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MUJ
 
 | | BT0996 RGII Chain B Complex | | Descriptor: | ACETATE ION, Beta-galactosidase, alpha-L-rhamnopyranose-(1-2)-[alpha-L-rhamnopyranose-(1-3)]alpha-L-arabinopyranose-(1-4)-[4-O-[(1R)-1-hydroxyethyl]-2-O-methyl-alpha-L-fucopyranose-(1-2)]beta-D-galactopyranose-(1-2)-alpha-D-aceric acid-(1-4)-alpha-L-rhamnopyranose-(1-3)-3-C-(hydroxylmethyl)-alpha-D-erythrofuranose | | Authors: | Cartmell, A, Basle, A, Ndeh, D, Luis, A.S, Venditto, I, Labourel, A, Rogowski, A, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQM
 
 | | Glycoside hydrolase BT_0986 | | Descriptor: | CALCIUM ION, D-rhamnopyranose tetrazole, Glycosyl hydrolases family 2, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQN
 
 | | Glycoside hydrolase BT_0986 | | Descriptor: | CALCIUM ION, Glycosyl hydrolases family 2, sugar binding domain | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MWK
 
 | | Glycoside hydrolase BT_0986 | | Descriptor: | BROMIDE ION, CALCIUM ION, Glycoside hydrolase family 2, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MSX
 
 | | Glycoside hydrolase BT_3662 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MUI
 
 | | Glycoside hydrolase BT_0996 | | Descriptor: | Beta-galactosidase, beta-L-arabinofuranose-(1-2)-alpha-L-rhamnopyranose-(1-2)-[alpha-L-rhamnopyranose-(1-3)]alpha-L-arabinopyranose-(1-4)-[4-O-[(1R)-1-hydroxyethyl]-2-O-methyl-alpha-L-fucopyranose-(1-2)]beta-D-galactopyranose-(1-2)-alpha-D-aceric acid-(1-3)-alpha-L-rhamnopyranose | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5G56
 
 | | THE TETRA-MODULAR CELLULOSOMAL ARABINOXYLANASE CtXyl5A STRUCTURE AS REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6 | | Authors: | Bras, J.L.A, Gilbert, H.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2016-05-21 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Mechanism by which Arabinoxylanases Can Recognise Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5LA1
 
 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5LA2
 
 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-L-arabinofuranose-(1-3)]alpha-D-xylopyranose, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5MQP
 
 | | Glycoside hydrolase BT_1002 | | Descriptor: | CALCIUM ION, Glycoside hydrolase BT_1002 | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The most complex carbohydrate known is degraded in the human gut by single organisms and not bacterial consortia
To Be Published
|
|
