6U5C
 
 | | RT XFEL structure of CypA solved using MESH injection system | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Wolff, A.M, Thompson, M.C. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Iucrj, 7, 2020
|
|
6U5E
 
 | | RT XFEL structure of CypA solved using celloluse carrier media | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Wolff, A.M, Nango, E, Nakane, T, Young, I.D, Brewster, A.S, Sugahara, M, Tanaka, R, Sauter, N.K, Tono, K, Iwata, S, Fraser, J.S, Thompson, M.C. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Iucrj, 7, 2020
|
|
6U5G
 
 | | MicroED structure of a FIB-milled CypA Crystal | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Wolff, A.M, Martynowycz, M.W, Zhao, W, Gonen, T, Fraser, J.S, Thompson, M.C. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Iucrj, 7, 2020
|
|
6U5D
 
 | | RT XFEL structure of CypA solved using LCP injection system | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Wolff, A.M, Young, I.D, Sierra, R.G, Brewster, A.S, Koralek, J.D, Boutet, S, Sauter, N.K, Fraser, J.S, Thompson, M.C. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Iucrj, 7, 2020
|
|
4Q0E
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-L-glutamic acid. | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, N-{4-[(2-amino-4-hydroxy-7H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzoyl}-L-glutamic acid, ... | | Authors: | Kumar, V.P, Anderson, K.S. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural studies provide clues for analog design of specific inhibitors of Cryptosporidium hominis thymidylate synthase-dihydrofolate reductase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4Q0D
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP, methotrexate and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-L-glutamic acid. | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, ... | | Authors: | Kumar, V.P, Anderson, K.S. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.449 Å) | | Cite: | Structural studies provide clues for analog design of specific inhibitors of Cryptosporidium hominis thymidylate synthase-dihydrofolate reductase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5UQE
 
 | | Multidomain structure of human kidney-type glutaminase(KGA/GLS) | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Pasquali, C.C, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2017-02-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5OQA
 
 | |
5OOZ
 
 | |
5OQE
 
 | |
5OQ9
 
 | |
7SUT
 
 | | Light harvesting phycobiliprotein HaPE645 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | (15,16)-DIHYDROBILIVERDIN (SINGLY LINKED), 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-18 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
1E9R
 
 | | Bacterial conjugative coupling protein TrwBdeltaN70. Trigonal form in complex with sulphate. | | Descriptor: | CONJUGAL TRANSFER PROTEIN TRWB, SULFATE ION | | Authors: | Gomis-Rueth, F.X, Moncalian, G, Cabezon, E, de la Cruz, F, Coll, M. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1E9S
 
 | | Bacterial conjugative coupling protein TrwBdeltaN70. Unbound monoclinic form. | | Descriptor: | CONJUGAL TRANSFER PROTEIN TRWB | | Authors: | Gomis-Rueth, F.X, Moncalian, G, Cabezon, E, de la Cruz, F, Coll, M. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
6RYA
 
 | | Structure of Dup1 mutant H67A:Ubiquitin complex | | Descriptor: | Polyubiquitin-C, Septation initiation protein | | Authors: | Donghyuk, S, Ivan, D. | | Deposit date: | 2019-06-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Regulation of Phosphoribosyl-Linked Serine Ubiquitination by Deubiquitinases DupA and DupB.
Mol.Cell, 77, 2020
|
|
6RYB
 
 | |
6ZQH
 
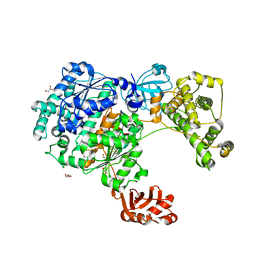 | | Yeast Uba1 in complex with ubiquitin | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Development of ADPribosyl Ubiquitin Analogues to Study Enzymes Involved in Legionella Infection.
Chemistry, 27, 2021
|
|
2V04
 
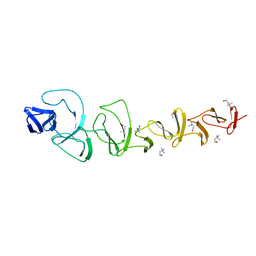 | | CRYSTAL STRUCTURE OF CHOLINE BINDING PROTEIN F FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CHOLINE BINDING PROTEIN F, CHOLINE ION | | Authors: | Hermoso, J, Molina, R, Kahn, R, Stelter, M. | | Deposit date: | 2007-05-08 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cbpf, a Bifunctional Choline-Binding Protein and Autolysis Regulator from Streptococcus Pneumoniae.
Embo Rep., 10, 2009
|
|
2V05
 
 | |
4DE7
 
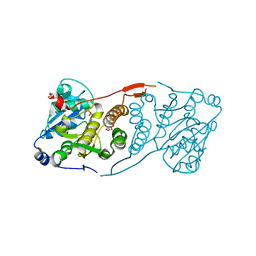 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mg2+ and uridine-diphosphate (UDP) | | Descriptor: | GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL, URIDINE-5'-DIPHOSPHATE | | Authors: | Albesa-Jove, D, Urresti, S, van der Woerd, M, Guerin, M.E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
4DDZ
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis | | Descriptor: | GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL | | Authors: | Albesa-Jove, D, Urresti, S, Gest, P.M, van der Woerd, M, Jackson, M, Guerin, M.E. | | Deposit date: | 2012-01-19 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
4DEC
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate (UDP) and phosphoglyceric acid (PGA) | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL, ... | | Authors: | Albesa-Jove, D, Urresti, S, van der Woerd, M, Guerin, M.E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
2HR6
 
 | | Crystal structure of dUTPase in complex with substrate analogue dUDP and manganese | | Descriptor: | 1,2-ETHANEDIOL, DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Kovari, J, Tapai, R, Vertessy, B.G. | | Deposit date: | 2006-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Methylene substitution at the alpha-beta bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site.
Proteins, 71, 2008
|
|
2HRM
 
 | | Crystal structure of dUTPase complexed with substrate analogue methylene-dUTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-5'-O-[(S)-HYDROXY(PHOSPHONOMETHYL)PHOSPHORYL]URIDINE, Deoxyuridine 5'-triphosphate nucleotidohydrolase | | Authors: | Barabas, O, Kovari, J, Tapai, R, Vertessy, B.G. | | Deposit date: | 2006-07-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Methylene substitution at the alpha-beta bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site.
Proteins, 71, 2008
|
|
1K3H
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
